Supplementary Table 2. ESNP-And ECNP-Related Genes List
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table 2. Functional Classification of Genes Differentially Regulated After HOXB4 Inactivation in HSC/Hpcs
Table 2. Functional classification of genes differentially regulated after HOXB4 inactivation in HSC/HPCs Symbol Gene description Fold-change (mean ± SD) Signal transduction Adam8 A disintegrin and metalloprotease domain 8 1.91 ± 0.51 Arl4 ADP-ribosylation factor-like 4 - 1.80 ± 0.40 Dusp6 Dual specificity phosphatase 6 (Mkp3) - 2.30 ± 0.46 Ksr1 Kinase suppressor of ras 1 1.92 ± 0.42 Lyst Lysosomal trafficking regulator 1.89 ± 0.34 Mapk1ip1 Mitogen activated protein kinase 1 interacting protein 1 1.84 ± 0.22 Narf* Nuclear prelamin A recognition factor 2.12 ± 0.04 Plekha2 Pleckstrin homology domain-containing. family A. (phosphoinosite 2.15 ± 0.22 binding specific) member 2 Ptp4a2 Protein tyrosine phosphatase 4a2 - 2.04 ± 0.94 Rasa2* RAS p21 activator protein 2 - 2.80 ± 0.13 Rassf4 RAS association (RalGDS/AF-6) domain family 4 3.44 ± 2.56 Rgs18 Regulator of G-protein signaling - 1.93 ± 0.57 Rrad Ras-related associated with diabetes 1.81 ± 0.73 Sh3kbp1 SH3 domain kinase bindings protein 1 - 2.19 ± 0.53 Senp2 SUMO/sentrin specific protease 2 - 1.97 ± 0.49 Socs2 Suppressor of cytokine signaling 2 - 2.82 ± 0.85 Socs5 Suppressor of cytokine signaling 5 2.13 ± 0.08 Socs6 Suppressor of cytokine signaling 6 - 2.18 ± 0.38 Spry1 Sprouty 1 - 2.69 ± 0.19 Sos1 Son of sevenless homolog 1 (Drosophila) 2.16 ± 0.71 Ywhag 3-monooxygenase/tryptophan 5- monooxygenase activation protein. - 2.37 ± 1.42 gamma polypeptide Zfyve21 Zinc finger. FYVE domain containing 21 1.93 ± 0.57 Ligands and receptors Bambi BMP and activin membrane-bound inhibitor - 2.94 ± 0.62 -
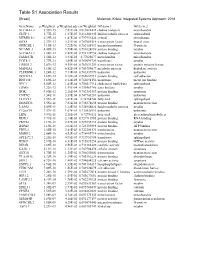
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

Functional Roles of Bromodomain Proteins in Cancer
cancers Review Functional Roles of Bromodomain Proteins in Cancer Samuel P. Boyson 1,2, Cong Gao 3, Kathleen Quinn 2,3, Joseph Boyd 3, Hana Paculova 3 , Seth Frietze 3,4,* and Karen C. Glass 1,2,4,* 1 Department of Pharmaceutical Sciences, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA; [email protected] 2 Department of Pharmacology, Larner College of Medicine, University of Vermont, Burlington, VT 05405, USA; [email protected] 3 Department of Biomedical and Health Sciences, University of Vermont, Burlington, VT 05405, USA; [email protected] (C.G.); [email protected] (J.B.); [email protected] (H.P.) 4 University of Vermont Cancer Center, Burlington, VT 05405, USA * Correspondence: [email protected] (S.F.); [email protected] (K.C.G.) Simple Summary: This review provides an in depth analysis of the role of bromodomain-containing proteins in cancer development. As readers of acetylated lysine on nucleosomal histones, bromod- omain proteins are poised to activate gene expression, and often promote cancer progression. We examined changes in gene expression patterns that are observed in bromodomain-containing proteins and associated with specific cancer types. We also mapped the protein–protein interaction network for the human bromodomain-containing proteins, discuss the cellular roles of these epigenetic regu- lators as part of nine different functional groups, and identify bromodomain-specific mechanisms in cancer development. Lastly, we summarize emerging strategies to target bromodomain proteins in cancer therapy, including those that may be essential for overcoming resistance. Overall, this review provides a timely discussion of the different mechanisms of bromodomain-containing pro- Citation: Boyson, S.P.; Gao, C.; teins in cancer, and an updated assessment of their utility as a therapeutic target for a variety of Quinn, K.; Boyd, J.; Paculova, H.; cancer subtypes. -

Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin
cells Review Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin Agnieszka Bochy ´nska,Juliane Lüscher-Firzlaff and Bernhard Lüscher * ID Institute of Biochemistry and Molecular Biology, Medical School, RWTH Aachen University, Pauwelsstrasse 30, 52057 Aachen, Germany; [email protected] (A.B.); jluescher-fi[email protected] (J.L.-F.) * Correspondence: [email protected]; Tel.: +49-241-8088850; Fax: +49-241-8082427 Received: 18 January 2018; Accepted: 27 February 2018; Published: 2 March 2018 Abstract: Regulation of gene expression is achieved by sequence-specific transcriptional regulators, which convey the information that is contained in the sequence of DNA into RNA polymerase activity. This is achieved by the recruitment of transcriptional co-factors. One of the consequences of co-factor recruitment is the control of specific properties of nucleosomes, the basic units of chromatin, and their protein components, the core histones. The main principles are to regulate the position and the characteristics of nucleosomes. The latter includes modulating the composition of core histones and their variants that are integrated into nucleosomes, and the post-translational modification of these histones referred to as histone marks. One of these marks is the methylation of lysine 4 of the core histone H3 (H3K4). While mono-methylation of H3K4 (H3K4me1) is located preferentially at active enhancers, tri-methylation (H3K4me3) is a mark found at open and potentially active promoters. Thus, H3K4 methylation is typically associated with gene transcription. The class 2 lysine methyltransferases (KMTs) are the main enzymes that methylate H3K4. KMT2 enzymes function in complexes that contain a necessary core complex composed of WDR5, RBBP5, ASH2L, and DPY30, the so-called WRAD complex. -

Histone-Binding of DPF2 Mediates Its Repressive Role in Myeloid Differentiation
Histone-binding of DPF2 mediates its repressive role in myeloid differentiation Ferdinand M. Hubera,1, Sarah M. Greenblattb,1, Andrew M. Davenporta,1, Concepcion Martinezb,YeXub,LyP.Vuc, Stephen D. Nimerb,2, and André Hoelza,2 aDivision of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125; bSylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL 33136; and cMolecular Pharmacology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065 Edited by Douglas C. Rees, Howard Hughes Medical Institute, California Institute of Technology, Pasadena, CA, and approved April 26, 2017 (received for review January 6, 2017) Double plant homeodomain finger 2 (DPF2) is a highly evolution- RUNX1 form a methylation-dependent repressive complex in arily conserved member of the d4 protein family that is ubiqui- AML, although it remains unclear whether the two proteins bind tously expressed in human tissues and was recently shown to each other directly or act concertedly as part of a larger complex. inhibit the myeloid differentiation of hematopoietic stem/progen- Here, we present the crystal structure of the human DPF2 itor and acute myelogenous leukemia cells. Here, we present the tandem PHD finger domain at a 1.6-Å resolution. We demon- crystal structure of the tandem plant homeodomain finger domain strate that the DPF2 tandem PHD finger domain binds acetylated of human DPF2 at 1.6-Å resolution. We show that DPF2 interacts H3 and H4 histone tails, identify the primary determinants of with the acetylated tails of both histones 3 and 4 via bipartite histone recognition, and confirm these interactions in vivo. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genome-Wide Rnai Screens Identify Genes Required for Ricin and PE Intoxications
Developmental Cell Article Genome-Wide RNAi Screens Identify Genes Required for Ricin and PE Intoxications Dimitri Moreau,1 Pankaj Kumar,1 Shyi Chyi Wang,1 Alexandre Chaumet,1 Shin Yi Chew,1 He´ le` ne Chevalley,1 and Fre´ de´ ric Bard1,* 1Institute of Molecular and Cell Biology, 61 Biopolis Drive, Proteos, Singapore 138673, Singapore *Correspondence: [email protected] DOI 10.1016/j.devcel.2011.06.014 SUMMARY In the lumen of the ER, these toxins are thought to interact with elements of the ER-associated degradation (ERAD) pathway, Protein toxins such as Ricin and Pseudomonas which targets misfolded proteins in the ER for degradation. exotoxin (PE) pose major public health challenges. This interaction is proposed to allow translocation to the cytosol Both toxins depend on host cell machinery for inter- without resulting in toxin degradation (Johannes and Ro¨ mer, nalization, retrograde trafficking from endosomes 2010). to the ER, and translocation to cytosol. Although Obviously, this complex set of membrane-trafficking and both toxins follow a similar intracellular route, it is membrane-translocation events involves many host proteins, some of which have already been described (Johannes and unknown how much they rely on the same genes. Ro¨ mer, 2010; Sandvig et al., 2010). Altering the function of these Here we conducted two genome-wide RNAi screens host proteins could in theory provide a toxin antidote. identifying genes required for intoxication and Consistently, inhibition of retrograde traffic by drugs such as demonstrating that requirements are strikingly Brefeldin A (Sandvig et al., 1991)(Yoshida et al., 1991) or Golgi- different between PE and Ricin, with only 13% over- cide A (Sa´ enz et al., 2009) and Retro-1 and 2 (Stechmann et al., lap. -

Epigenetics Page 1
Epigenetics esiRNA ID Gene Name Gene Description Ensembl ID HU-13237-1 ACTL6A actin-like 6A ENSG00000136518 HU-13925-1 ACTL6B actin-like 6B ENSG00000077080 HU-14457-1 ACTR1A ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) ENSG00000138107 HU-10579-1 ACTR2 ARP2 actin-related protein 2 homolog (yeast) ENSG00000138071 HU-10837-1 ACTR3 ARP3 actin-related protein 3 homolog (yeast) ENSG00000115091 HU-09776-1 ACTR5 ARP5 actin-related protein 5 homolog (yeast) ENSG00000101442 HU-00773-1 ACTR6 ARP6 actin-related protein 6 homolog (yeast) ENSG00000075089 HU-07176-1 ACTR8 ARP8 actin-related protein 8 homolog (yeast) ENSG00000113812 HU-09411-1 AHCTF1 AT hook containing transcription factor 1 ENSG00000153207 HU-15150-1 AIRE autoimmune regulator ENSG00000160224 HU-12332-1 AKAP1 A kinase (PRKA) anchor protein 1 ENSG00000121057 HU-04065-1 ALG13 asparagine-linked glycosylation 13 homolog (S. cerevisiae) ENSG00000101901 HU-13552-1 ALKBH1 alkB, alkylation repair homolog 1 (E. coli) ENSG00000100601 HU-06662-1 ARID1A AT rich interactive domain 1A (SWI-like) ENSG00000117713 HU-12790-1 ARID1B AT rich interactive domain 1B (SWI1-like) ENSG00000049618 HU-09415-1 ARID2 AT rich interactive domain 2 (ARID, RFX-like) ENSG00000189079 HU-03890-1 ARID3A AT rich interactive domain 3A (BRIGHT-like) ENSG00000116017 HU-14677-1 ARID3B AT rich interactive domain 3B (BRIGHT-like) ENSG00000179361 HU-14203-1 ARID3C AT rich interactive domain 3C (BRIGHT-like) ENSG00000205143 HU-09104-1 ARID4A AT rich interactive domain 4A (RBP1-like) ENSG00000032219 HU-12512-1 ARID4B AT rich interactive domain 4B (RBP1-like) ENSG00000054267 HU-12520-1 ARID5A AT rich interactive domain 5A (MRF1-like) ENSG00000196843 HU-06595-1 ARID5B AT rich interactive domain 5B (MRF1-like) ENSG00000150347 HU-00556-1 ASF1A ASF1 anti-silencing function 1 homolog A (S. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Novel Driver Strength Index Highlights Important Cancer Genes in TCGA Pancanatlas Patients
medRxiv preprint doi: https://doi.org/10.1101/2021.08.01.21261447; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Novel Driver Strength Index highlights important cancer genes in TCGA PanCanAtlas patients Aleksey V. Belikov*, Danila V. Otnyukov, Alexey D. Vyatkin and Sergey V. Leonov Laboratory of Innovative Medicine, School of Biological and Medical Physics, Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia *Corresponding author: [email protected] NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. 1 medRxiv preprint doi: https://doi.org/10.1101/2021.08.01.21261447; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Abstract Elucidating crucial driver genes is paramount for understanding the cancer origins and mechanisms of progression, as well as selecting targets for molecular therapy. Cancer genes are usually ranked by the frequency of mutation, which, however, does not necessarily reflect their driver strength. Here we hypothesize that driver strength is higher for genes that are preferentially mutated in patients with few driver mutations overall, because these few mutations should be strong enough to initiate cancer. -

Antigen-Specific Memory CD4 T Cells Coordinated Changes in DNA
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology The Journal of Immunology published online 18 March 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/03/17/jimmun ol.1202267 Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto, Katsumi Ogoshi, Atsushi Sasaki, Jun Abe, Wei Qu, Yoichiro Nakatani, Budrul Ahsan, Kenshiro Oshima, Francis H. W. Shand, Akio Ametani, Yutaka Suzuki, Shuichi Kaneko, Takashi Wada, Masahira Hattori, Sumio Sugano, Shinichi Morishita and Kouji Matsushima J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Author Choice option Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Freely available online through http://www.jimmunol.org/content/suppl/2013/03/18/jimmunol.120226 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Author Choice Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. Published March 18, 2013, doi:10.4049/jimmunol.1202267 The Journal of Immunology Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto,*,†,‡ Katsumi Ogoshi,* Atsushi Sasaki,† Jun Abe,* Wei Qu,† Yoichiro Nakatani,† Budrul Ahsan,x Kenshiro Oshima,† Francis H. -

Regular Article
From www.bloodjournal.org by guest on April 6, 2015. For personal use only. Regular Article IMMUNOBIOLOGY Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion Waldo A. Spessott,1 Maria L. Sanmillan,1 Margaret E. McCormick,1 Nishant Patel,2 Joyce Villanueva,3 Kejian Zhang,4 Kim E. Nichols,5 and Claudio G. Giraudo1 1Department of Pathology and Laboratory Medicine, and 2Division of Oncology, Department of Pediatrics, The Children’s Hospital of Philadelphia, University of Pennsylvania, Philadelphia, PA; 3Division of Bone Marrow Transplant and Immune Deficiency, and 4Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH; and 5Division of Cancer Predisposition, Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN Key Points Familial hemophagocytic lymphohistiocytosis (F-HLH) and Griscelli syndrome type 2 (GS) are life-threatening immunodeficiencies characterized by impaired cytotoxic T lymphocyte • Monoallelic STXBP2 mutations (CTL) and natural killer (NK) cell lytic activity. In the majority of cases, these disorders are affecting codon 65 impair caused by biallelic inactivating germline mutations in genes such as RAB27A (GS) and PRF1, lymphocyte cytotoxicity and UNC13D, STX11,andSTXBP2 (F-HLH). Although monoallelic (ie, heterozygous) mutations contribute to hemophagocytic have been identified in certain patients, the clinical significance and molecular mechanisms lymphohistiocytosis. by which these mutations influence CTL and NK cell function remain poorly understood. • Munc18-2R65Q/W mutant Here, we characterize 2 novel monoallelic hemophagocytic lymphohistiocytosis (HLH)- associated mutations affecting codon 65 of STXPB2, the gene encoding Munc18-2, a member proteins function in a dominant- of the SEC/MUNC18 family.