Members of the Competence Network for Congenital Heart Defects, Germany
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Influencers on Thyroid Cancer Onset: Molecular Genetic Basis
G C A T T A C G G C A T genes Review Influencers on Thyroid Cancer Onset: Molecular Genetic Basis Berta Luzón-Toro 1,2, Raquel María Fernández 1,2, Leticia Villalba-Benito 1,2, Ana Torroglosa 1,2, Guillermo Antiñolo 1,2 and Salud Borrego 1,2,* 1 Department of Maternofetal Medicine, Genetics and Reproduction, Institute of Biomedicine of Seville (IBIS), University Hospital Virgen del Rocío/CSIC/University of Seville, 41013 Seville, Spain; [email protected] (B.L.-T.); [email protected] (R.M.F.); [email protected] (L.V.-B.); [email protected] (A.T.); [email protected] (G.A.) 2 Centre for Biomedical Network Research on Rare Diseases (CIBERER), 41013 Seville, Spain * Correspondence: [email protected]; Tel.: +34-955-012641 Received: 3 September 2019; Accepted: 6 November 2019; Published: 8 November 2019 Abstract: Thyroid cancer, a cancerous tumor or growth located within the thyroid gland, is the most common endocrine cancer. It is one of the few cancers whereby incidence rates have increased in recent years. It occurs in all age groups, from children through to seniors. Most studies are focused on dissecting its genetic basis, since our current knowledge of the genetic background of the different forms of thyroid cancer is far from complete, which poses a challenge for diagnosis and prognosis of the disease. In this review, we describe prevailing advances and update our understanding of the molecular genetics of thyroid cancer, focusing on the main genes related with the pathology, including the different noncoding RNAs associated with the disease. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Molecular Mechanisms Underlying Innate Immune Kinase
MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION APPROVED BY SUPERVISORY COMMITTEE Michael A White, Ph.D. Melanie H. Cobb, Ph.D. Lawrence Lum, Ph.D. John D. Minna, M.D. DEDICATION This work is dedicated to my mother and Arlene for their love and support. ACKNOWLEDGEMENTS I am very grateful to my mentor, Dr. Michael White, for his continuous support and guidance through the entire study. I really appreciate his inspiration, patience, and generosity. I would also like to thank my committee members, Dr. Cobb, Dr. Lum, and Dr. Minna, for their invaluable advice and discussion. I thank all the White lab members and my friends for their help, suggestion, and discussion. Particularly, I would like to thank Rosie, Michael, Brian, Tzuling, and Malia for their long-term support and collaboration. I would also like to thank my friends, Veleka, Pei-Ling, Jen-Chieh, Shu-Yi, Chih-Chiang, Jen-Shuan, Yi-Chun, and Yu-San for their friendship. I am grateful to Drs. Rolf Brekken, Zhijian James Chen, Xuetao Cao, Philip Tsichlis, Charles Yeaman, William Hahn, Keqiang Ye, and Shu-Chan Hsu, Bing Su, Dos Sarbassov, Mark Magnuson, David Sabatini, Thomas Tan, and Bert Vogelstein for many of the reagents used in these studies. Finally, and most importantly, I would like to thank my mother and Arlene for their unending support and encouragement. MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION by YI-HUNG OU DISSERTATION Presented to the Faculty of the Graduate School of Biomedical Sciences The University of Texas Southwestern Medical Center at Dallas In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY The University of Texas Southwestern Medical Center at Dallas Dallas, Texas April, 2013 Copyright by YI-HUNG OU, 2013 All Rights Reserved MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION Publication No. -
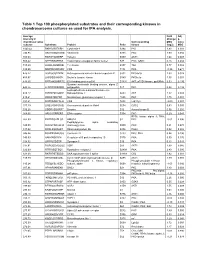
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5. -

The Curing AI for Precision Medicine
The Curing AI for Precision Medicine Hoifung Poon 1 Medicine Today Is Imprecise Top 20 drugs 80% non-responders Wasted 1/3 health spending $750 billion / year 2 Disruption 1: Big Data 2009 2013: 40% 93% 3 Disruption 2: Pay-for-Performance Goal: 75% by 2020 4 Vemurafenib on BRAF-V600 Melanoma Before Treatment 15 Weeks 5 Vemurafenib on BRAF-V600 Melanoma Before Treatment 15 Weeks 23 Weeks 6 Why We Haven’t Solved Precision Medicine? … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … High-Throughput Data Discovery Bottleneck #1: Knowledge Bottleneck #2: Reasoning AI is the key to overcome these bottlenecks 7 Use Case: Molecular Tumor Board 8 www.ucsf.edu/news/2014/11/120451/bridging-gap-precision-medicine Use Case: Molecular Tumor Board Problem: Hard to scale U.S. 2015: 1.6 million new cases, 600K deaths 902 cancer hospitals Memorial Sloan Kettering 2016: Sequence: Tens of thousand Board can review: A few hundred Wanted: Decision support for cancer precision medicine 9 First-Generation Molecular Tumor Board Knowledge bottleneck E.g., given a tumor sequence, determine: What genes and mutations are important What drugs might be applicable Can do manually but hard to scale 10 Next-Generation Molecular Tumor Board Reasoning bottleneck E.g., personalize drug combinations Can’t do manually, ever 11 Big Medical Data Decision Support Precision Medicine Machine Predict Reading Drug Combo 12 13 PubMed 26 millions abstracts Two new abstracts every minute Adds over one million every year 14 Machine Reading PMID: 123 … VDR+ binds to SMAD3 to form … PMID: 456 Knowledge … JUN expression Base is induced by SMAD3/4 … …… 15 Machine Reading Involvement of p70(S6)-kinase activation in IL-10 up-regulation in human monocytes by gp41 envelope protein of human immunodeficiency virus type 1 .. -

Cross‑Talks Between Micrornas and Mrnas in Pancreatic Tissues of Streptozotocin‑Induced Type 1 Diabetic Mice
BIOMEDICAL REPORTS 3: 333-342, 2015 Cross‑talks between microRNAs and mRNAs in pancreatic tissues of streptozotocin‑induced type 1 diabetic mice CAIMING TIAN1,2*, XIAOXI OUYANG3*, QING LV1,4, YAOU ZHANG2 and WEIDONG XIE2 1Department of Chemistry, Tsinghua University, Beijing 100084; 2Shenzhen Key Laboratory of Health Science and Technology, Division of Life Science and Health, Graduate School at Shenzhen, Tsinghua University, Shenzhen, Guangdong 518055; 3Department of Health Inspection and Quarantine, School of Public Health, Sun Yat-sen University, Guangzhou, Guangdong 510080; 4School of Life Sciences, Tsinghua University, Beijing 100084, P.R. China Received December 30, 2014; Accepted February 2, 2015 DOI: 10.3892/br.2015.426 Abstract. Network cross-talks between microRNAs (miRNAs) Introduction and mRNAs may be useful to elucidate the pathological mechanisms of pancreatic islet cells in diabetic individuals. Diabetes mellitus is a complex metabolic disease demon- The aim of the present study was to investigate the cross-talks strating impaired islet function (1,2). Destruction of β-cells between miRNAs and mRNAs in pancreatic tissues of or the failure of these insulin-secreting cells to compensate streptozotocin-induced diabetic mice through microarray and for increased metabolic demand may account for the develop- bioinformatic methods. Based on the miRNA microarray, ment of diabetes. Apoptosis, oxidative stress, mitochondrial 64 upregulated and 72 downregulated miRNAs were observed dysfunction and endoplasmic reticulum (ER) stress responses, in pancreatic tissues in diabetic mice compared to the normal including c-Jun N-terminal kinase (JNK) activation, have been controls. Based on the mRNA microarrray, 507 upregulated suggested as mechanisms for the changes of pancreatic β-cells mRNAs and 570 downregulated mRNAs were identified in in type 2 diabetes mellitus (T2DM) (3). -

A Microgravity Environment Improves Structural Resolution and Endows Cues for Specific Inhibition of Mitogen-Activated Protein Kinase Kinase 7
Int. J. Microgravity Sci. Appl., 36 (1) (2019) 360102 DOI: 10.15011//jasma.36.360102 IIII Original Paper IIII A Microgravity Environment Improves Structural Resolution and Endows Cues for Specific Inhibition of Mitogen-Activated Protein Kinase Kinase 7 Takayoshi KINOSHITA1, Takuma HASHIMOTO1, Yuka MURAKAWA1, Yuri SOGABE1, 2 3 Takashi MATSUMOTO and Masaaki SAWA Abstract Mitogen-activated protein kinase kinase 7 (MAP2K7) regulates stress and inflammatory responses, and is an attractive drug discovery target for serious diseases such as arthritis and cardiac hypertrophy. A microgravity environment improved the crystal quality of MAP2K7 and improved the structural resolution to 1.3 Å. High resolution analysis structurally clarified the two regions, which were undefined in the previous low-resolution analysis and conferred structural insights for producing MAP2K7-specific inhibitors. The hinge region alternatively configures the canonical and atypical conformations. The latter could allow binding MAP2K7 inhibitors with a novel scaffold. The C-terminal region works as a negative regulator with intermolecular association, which implies the ability to produce highly selective MAP2K7 inhibitors. Keyword(s): MAP2K7, High-resolution structure, Crystallization in microgravity, Allosteric regulation, Specific inhibition. Received 30 October 2018, Accepted 19 December 2018, Published 31 January 2019. 1. Introduction paper, we will dissect this structure using the additional biochemical data and depict the crucial cues for producing Mitogen-activated protein kinase kinase 7 (MAP2K7) plays a MAP2K7-specific inhibitors. crucial role in the c-Jun N-terminal kinase (JNK) signaling cascade, which regulates stress and inflammatory responses 1), 2. Experimental and is an attractive drug discovery target for serious diseases, including arthritis, hepatoma, and cardiac hypertrophy 2–4). -

Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM 014911.2 KIAA1048, Dkfzp686k16132 AP2 Associated Kinase 1
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAK1 NM_014911.2 KIAA1048, DKFZp686K16132 AP2 associated kinase 1 (AAK1) AATK NM_001080395.2 AATYK, AATYK1, KIAA0641, LMR1, LMTK1, p35BP apoptosis-associated tyrosine kinase (AATK) ABL1 NM_007313.2 ABL, JTK7, c-ABL, p150 v-abl Abelson murine leukemia viral oncogene homolog 1 (ABL1) ABL2 NM_007314.3 ABLL, ARG v-abl Abelson murine leukemia viral oncogene homolog 2 (arg, Abelson-related gene) (ABL2) ACVR1 NM_001105.2 ACVRLK2, SKR1, ALK2, ACVR1A activin A receptor ACVR1B NM_004302.3 ACVRLK4, ALK4, SKR2, ActRIB activin A receptor, type IB (ACVR1B) ACVR1C NM_145259.2 ACVRLK7, ALK7 activin A receptor, type IC (ACVR1C) ACVR2A NM_001616.3 ACVR2, ACTRII activin A receptor ACVR2B NM_001106.2 ActR-IIB activin A receptor ACVRL1 NM_000020.1 ACVRLK1, ORW2, HHT2, ALK1, HHT activin A receptor type II-like 1 (ACVRL1) ADCK1 NM_020421.2 FLJ39600 aarF domain containing kinase 1 (ADCK1) ADCK2 NM_052853.3 MGC20727 aarF domain containing kinase 2 (ADCK2) ADCK3 NM_020247.3 CABC1, COQ8, SCAR9 chaperone, ABC1 activity of bc1 complex like (S. pombe) (CABC1) ADCK4 NM_024876.3 aarF domain containing kinase 4 (ADCK4) ADCK5 NM_174922.3 FLJ35454 aarF domain containing kinase 5 (ADCK5) ADRBK1 NM_001619.2 GRK2, BARK1 adrenergic, beta, receptor kinase 1 (ADRBK1) ADRBK2 NM_005160.2 GRK3, BARK2 adrenergic, beta, receptor kinase 2 (ADRBK2) AKT1 NM_001014431.1 RAC, PKB, PRKBA, AKT v-akt murine thymoma viral oncogene homolog 1 (AKT1) AKT2 NM_001626.2 v-akt murine thymoma viral oncogene homolog 2 (AKT2) AKT3 NM_181690.1 -

Rabbit Anti-Human MAP2K5 Polyclonal Antibody (DPABH-17348) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Rabbit Anti-Human MAP2K5 Polyclonal antibody (DPABH-17348) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Immunogen MEK5 fusion protein, sequence: GQLIEPLQIFPRACKPPGERNIHGLKVNTRAGPSQHSSPAVSDSLPSNSLKKSSAELKKILANGQ MNEQDIRYRDTLGHGNGGTVYKAYHVPSGKILAVKVILLDITLELQKQIMSELEILYKCDSSYIIGF YGAFFVENRISICTEFMDGGSLDVYRKMPEHVLGRIAVAVVKGLTYLWSLKILHRDVKPSNMLVN TRGQVKLCDFGVSTQLVNSIAKTYVGTNAYMAPERISGEQYGIHSDVWSLGISFMELALGRFPY PQIQKNQGSLMPLQLLQCIVDEDSPVLPVGEFSEPFVHFITQCMRKQPKERPAPEELMGHPFIV QFNDGNAAVVSMWVCRALEERRSQQGPP (96-448aa encoded by BC008838) Isotype IgG Source/Host Rabbit Species Reactivity Human, Mouse, Rat Purification Antigen affinity purification Conjugate Unconjugated Applications WB, IHC, IF, ELISA Positive Control HeLa cells, A431 cells, HepG2 cells Format Liquid Size 50 uL; 100 uL Buffer PBS with 0.02% sodium azide and 50% glycerol pH 7.3. Preservative 0.02% Sodium Azide Storage Store at -20°C. Aliquoting is unnecessary for -20°C storage. BACKGROUND Introduction The protein encoded by this gene is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase specifically interacts with and activates MAPK7/ERK5. This 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved kinase itself can be phosphorylated and activated by MAP3K3/MEKK3, as well as by atypical protein kinase C isoforms (aPKCs). The signal cascade mediated by this kinase is involved in growth -

Brafnon-V600E More Frequently Co-Occurs with IDH1/2 Mutation in Adult Patients with Gliomas Than Patients Harboring BRAFV600E, but Without Survival Advantage
BRAFnon-V600E more frequently co-occurs with IDH1/2 mutation in adult patients with gliomas than patients harboring BRAFV600E, but without survival advantage Rong Da The First Alated Hospital of Xi'an Jiaotong University Maode Wang The First Alated Hospital of Xi'an Jiaotong University Haitao Jiang The First Aliated Hospital of Xi'an Jiaotong University Tuo Wang The First Aliated Hospital of Xi'an Jiaotong University Wei Wang ( [email protected] ) The First Aliated Hospital of Xi'an Jiaotong University https://orcid.org/0000-0002-6954-9056 Research article Keywords: patient with glioma, BRAFnon-V600E, BRAFV600E, IDH1/2 Posted Date: March 2nd, 2021 DOI: https://doi.org/10.21203/rs.3.rs-279542/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/17 Abstract Background The effect of BRAFnon−V600E and BRAFV600E on the outcome and the molecular characteristics in adult glioma patients is unknown and needs to be explored, although BRAFV600E has been extensively studied in pediatric glioma. Methods Co-occurring mutations and copy number alterations of associated genes in the MAPK and p53 pathways were retrieved and investigated using the cBioPortal. The prognosis of available adult glioma cohorts with BRAFV600E and BRAFnon−V600E mutations was also investigated. Results Glioblastoma multiform was the most common cancer type with BRAF non−V600E and BRAFV600E. TP53 (56.00% vs. 7.41%), IDH1/2 (36.00% vs. 3.70%), and ATRX (32.00% vs. 7.41%) exhibited more mutations in BRAFnon−V600E than in BRAFV600E, and TP53 was the independent risk factor (56.00% vs.