Development of a Conjugate Vaccine Against SARS-Cov-2 Nathan Wymer, Phd (Department of Chemistry and Biochemistry); Lindsey M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Commonly Administered Vaccines
Commonly Administered Vaccines CPT CVX Vaccine Type Brand Name Manufacturer PhilaVax Display Name Description Code Code Combination Vaccines Diptheria, tetanus toxoids and acellular pertussis vaccine, Hepati- DTaP-HepB-IPV Pediarix® GlaxoSmithKline 90723 110 DTaP-HepB-IPV tis B and poliovirus vaccine, inactivated Diptheria, tetanus toxoids and acellular pertussis vaccine and DTaP-Hib TriHIBit® Sanofi Pasteur 90721 50 DTaP-Hib (TriHIBit) Haemophilus influenzae type b conjugate vaccine Diptheria, tetanus toxoids and acellular pertussis vaccine, Hae- DTaP-Hib-IPV Pentacel® Sanofi Pasteur 90698 120 DTaP-Hib-IPV mophilus influenzae type b, and poliovirus vaccine, inactivated Diptheria, tetanus toxoids and acellular pertussis vaccine, and DTaP-IPV Kinrix® GlaxoSmithKline 90696 130 DTaP-IPV (KINRIX) poliovirus vaccine, inactivated HepA-HepB TWINRIX® GlaxoSmithKline 90636 104 HepA/B (TWINRIX) Hepatisis A and Hepatitis B vaccine, adult dosage Hepatitis B and Hemophilus influenza b vaccine, for intramuscular HepB-Hib Comvax® Merck 90748 51 Hib-Hep B (COMVAX) use Haemophilus influenza b and meningococcal sero groups C and Y MeningC/Y-Hib Menhibrix® GlaxoSmithKline 90644 148 Meningococcal-Hib vaccine, 4 dose series Diphtheria, Tetanus and Pertussis DTaP Infanrix® GlaxoSmithKline 90700 20 DTaP (INFANRIX) Diptheria, tetanus toxoids and acellular pertussis vaccine DTaP, 5 Pertussis Antigens Daptacel® Sanofi Pasteur 90700 106 DTaP (DAPTACEL) Diptheria, tetanus toxoids and acellular pertussis vaccine Tetanus toxoid, reduced diphtheria toxoid, and acellular -

Vaccine Hesitancy
Vaccine Hesitancy Dr Brenda Corcoran National Immunisation Office Presentation Outline An understanding of the following principles: • Overview of immunity • Different types of vaccines and vaccine contents • Vaccine failures • Time intervals between vaccine doses • Vaccine overload • Adverse reactions • Herd immunity Immunity Immunity • The ability of the human body to protect itself from infectious disease The immune system • Cells with a protective function in the – bone marrow – thymus – lymphatic system of ducts and nodes – spleen –blood Types of immunity Source: http://en.wikipedia.org/wiki/Immunological_memory Natural (innate) immunity Non-specific mechanisms – Physical barriers • skin and mucous membranes – Chemical barriers • gastric and digestive enzymes – Cellular and protein secretions • phagocytes, macrophages, complement system ** No “memory” of protection exists afterwards ** Passive immunity – adaptive mechanisms Natural • maternal transfer of antibodies to infant via placenta Artificial • administration of pre- formed substance to provide immediate but short-term protection (antitoxin, antibodies) Protection is temporary and wanes with time (usually few months) Active immunity – adaptive mechanisms Natural • following contact with organism Artificial • administration of agent to stimulate immune response (immunisation) Acquired through contact with an micro-organism Protection produced by individual’s own immune system Protection often life-long but may need boosting How vaccines work • Induce active immunity – Immunity and immunologic memory similar to natural infection but without risk of disease • Immunological memory allows – Rapid recognition and response to pathogen – Prevent or modify effect of disease Live attenuated vaccines Weakened viruses /bacteria – Achieved by growing numerous generations in laboratory – Produces long lasting immune response after one or two doses – Stimulates immune system to react as it does to natural infection – Can cause mild form of the disease (e.g. -

Criteria for Reviewing Pneumococcal Conjugate Vaccine
Draft—3/11/10 Oregon School/Facility Immunization Advisory Committee: Review of Pneumococcal Conjugate Vaccine Against Twelve Criteria for Children’s Facility Immunization Requirements Oregon Department of Human Services Public Health Division Office of Family Health Immunization Program 800 NE Oregon Street, Suite 370 Portland, Oregon 97232 Phone: 971-673-0300 Fax: 971-673-0278 Web: www.oregon.gov/DHS/ph/imm Page 1–Review of Pneumococcal Conjugate Vaccine Against Criteria for School Law Immunization Requirements Oregon School/Facility Immunization Advisory Committee: Review of Pneumococcal Conjugate Vaccine Against Twelve Criteria for School/Facility/College Immunization Requirements Process for Reviewing Antigens for Potential Inclusion in OAR 333-050-0050, 333-050-0130 and 333-050-0140. Request for the inclusion of additional antigens or vaccines can come from the Oregon Immunization Program, IPAT (Immunization Policy Advisory Team), or from the community. Proposed changes to vaccine requirements are discussed with IPAT either in a regularly scheduled meeting or through electronic communication. IPAT will submit their comments and a request for consideration to the Oregon Immunization School Law Advisory Committee. The Oregon School/Facility Immunization Advisory Committee was established as a part of the school law immunization requirements when the original legislation was passed in 1980. This Committee is composed of immunization stakeholders from the fields of public health, school health, school administration, medicine, day care, child advocacy and consumers (parents). Through consensus, the committee determines what vaccines (antigens) should be included in Oregon school immunization requirements. Information about new vaccines and the diseases they prevent, including transmission within schools, burden of disease, cost-effectiveness, effect on schools/counties and vaccine availability is presented at a scheduled meeting for committee consideration. -

Immunity and How Vaccines Work
Immunity and how vaccines work Dr Brenda Corcoran National Immunisation Office Presentation Outline An understanding of the following principles: • Overview of immunity • Different types of vaccines and vaccine contents • Vaccine failures • Time intervals between vaccine doses • Vaccine overload • Adverse reactions • Herd immunity Immunity Immunity • The ability of the human body to protect itself from infectious disease The immune system • Cells with a protective function in the – bone marrow – thymus – lymphatic system of ducts and nodes – spleen –blood Types of immunity Source: http://en.wikipedia.org/wiki/Immunological_memory Natural (innate) immunity Non-specific mechanisms – Physical barriers • skin and mucous membranes – Chemical barriers • gastric and digestive enzymes – Cellular and protein secretions • phagocytes, macrophages, complement system ** No “memory” of protection exists afterwards ** Passive immunity – adaptive mechanisms Natural • maternal transfer of antibodies to infant via placenta Artificial • administration of pre- formed substance to provide immediate but short-term protection (antitoxin, antibodies) Protection is temporary and wanes with time (usually few months) Active immunity – adaptive mechanisms Natural • following contact with organism Artificial • administration of agent to stimulate immune response (immunisation) Acquired through contact with an micro-organism Protection produced by individual’s own immune system Protection often life-long but may need boosting How vaccines work • Induce active immunity – Immunity and immunologic memory similar to natural infection but without risk of disease • Immunological memory allows – Rapid recognition and response to pathogen – Prevent or modify effect of disease Live attenuated vaccines Weakened viruses /bacteria – Achieved by growing numerous generations in laboratory – Produces long lasting immune response after one or two doses – Stimulates immune system to react as it does to natural infection – Can cause mild form of the disease (e.g. -

2021 Medicare Vaccine Coverage Part B Vs Part D
CDPHP® Medicare Advantage Vaccine Coverage Guide Part B (Medical) vs. Part D (Pharmacy) Medicare Part B (Medical): Medicare Part D (Pharmacy): Vaccinations or inoculations Vaccinations or inoculations are included when the administration is (except influenza, pneumococcal, reasonable and necessary for the prevention of illness. and hepatitis B for members at risk) are excluded unless they are directly related to the treatment of an injury or direct exposure to a disease or condition. • Influenza Vaccine (Flu) • BCG Vaccine • Pneumococcal Vaccine • Diphtheria/Tetanus/Acellular Pertussis Vaccine (ADACEL, (Pneumovax, Prevnar 13) BOOSTRIX, DAPTACEL, INFANRIX) • Hepatitis B Vaccine • Diphtheria/Tetanus/Acellular Pertussis/Inactivated Poliovirus (Recombivax, Engerix-B) Vaccine (KINRIX, QUADRACEL) for members at moderate • Diphtheria/Tetanus Vaccine (DT, Td, TDVAX, TENIVAC) to high risk • Diphtheria/Tetanus/Acellular Pertussis/Inactivated Poliovirus Vac • Other vaccines when directly cine/ Haemophilus Influenzae Type B Conjugate Vaccine (PENTACEL) related to the treatment of an • Diphtheria/Tetanus/Acellular Pertussis/Inactivated Poliovirus injury or direct exposure to a Vaccine/Hepatitis B Vaccine (PEDIARIX) disease or condition, such as: • Haemophilus Influenzae Type B Conjugate Vaccine (ActHIB, PedvaxHIB, • Antivenom Sera Hiberix) • Diphtheria/Tetanus Vaccine • Hepatitis A Vaccine, Inactivated (VAQTA) (DT, Td, TDVAX, TENIVAC) • Hepatitis B Vaccine, Recombinant (ENGERIX-B, RECOMBIVAX HB) • Rabies Virus Vaccine for members at low risk (RabAvert, -
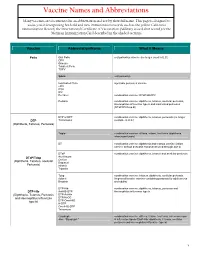
Vaccine Names and Abbreviations Vaccine Names and Abbreviations Many Vaccines Are Documented in an Abbreviation and Not by Their Full Name
Vaccine Names and Abbreviations Vaccine Names and Abbreviations Many vaccines are documented in an abbreviation and not by their full name. This page is designed to assist you in interpreting both old and new immunization records such as the yellow California Immunization Record, the International Certificate of Vaccination (Military issued shot record) or the Mexican Immunization Card described in the shaded sections. Vaccine Abbreviation/Name What it Means Polio Oral Polio oral poliovirus vaccine (no longer used in U.S.) OPV Orimune Trivalent Polio TOPV Sabin oral poliovirus Inactivated Polio injectable poliovirus vaccine eIPV IPOL IPV Pentacel combination vaccine: DTaP/Hib/IPV Pediarix combination vaccine: diphtheria, tetanus, acellular pertussis, Haemophilus influenzae type b and inactivated poliovirus (DTaP/IPV/Hep B) DTP or DPT combination vaccine: diphtheria, tetanus, pertussis (no longer DTP Tri-Immunol available in U.S.) (Diphtheria, Tetanus, Pertussis) Triple combination vaccine: difteria, tétano, tos ferina (diphtheria, tetanus pertussis) DT combination vaccine: diphtheria and tetanus vaccine (infant vaccine without pertussis component used through age 6) DTaP combination vaccine: diphtheria, tetanus and acellular pertussis DTaP/Tdap Acel-Imune Certiva (Diphtheria, Tetanus, acellular Daptacel Pertussis) Infanrix Tripedia Tdap combination vaccine: tetanus, diphtheria, acellular pertussis Adacel (improved booster vaccine containing pertussis for adolescents Boostrix and adults) DTP-Hib combination vaccine: diphtheria, tetanus, -

Vaccines and Immunization February 20-21, 2017 Berlin, Germany
Lallan Giri, J Vaccines Vaccin 2017, 8:1(Suppl) conferenceseries.com http://dx.doi.org/10.4172/2157-7560.C1.054 15th Annual Summit on Vaccines and Immunization February 20-21, 2017 Berlin, Germany Protection against bioterrorism Lallan Giri Biologics Resources LLC, USA ioterrorism involves the intentional release of dissemination of deadly biological agents in the cities and communities. Terrorism Bhas become a common and easier way of causing harms to innocent public. At present time, it has caused serious concerns and insecurity in the public domain. One of the sources or methods used by terrorists is through dissemination of deadly bacteria, viruses, and chemicals. Bioterrorism agents are found typically in nature and can be grown easily. It is also possible that they can be mutated or altered to increase their ability to cause deadly diseases causing mass casualty of human lives. Anthrax letter attacks after September 11, 2001 demonstrated the ease of using the anthrax spores as a weapon of mass destruction in a bioterrorist attack. Anthrax is lethal infectious disease caused by the spore forming Bacillus anthracis. The two major virulence factors of B. anthracis are the poly-y-glutamic acid (Y-D-PGA) capsule and exotoxin. The three components of the exotoxin, Protective Antigen (PA), Lethal Factor (LF) and Edema Factor (EF) are produced and secreted separately and form two bipartite toxins that cause massive edema and organ failure. The anti-phagocytic y-D-PGA capsule disguises the bacilli from immune surveillance by macrophages and allows unimpeded growth of the bacilli leading to anthrax disease caused by both, the capsule and toxins. -

Pneumococcal Conjugate Vaccine: Supply & Demand Update August 2016
Pneumococcal Conjugate Vaccine: Supply & Demand Update UNICEF Supply Division August 2016 0 Pneumococcal Conjugate Vaccine: Supply & Demand Update August 2016 This update provides new information on countries, scheduled pneumococcal conjugate vaccine (PCV) introductions, and supply availability. PCV supply availability through UNICEF remained stable during 2015, and is no longer constrained on account of increased production capacity. UNICEF will continue to assess the needs for future demand. A more recent note covering PCV exists. Please visit https://www.unicef.org/supply/market-notes- and-updates 1. Summary • During 2014 through end-July 2016, nineteen countries, supported by Gavi, the Vaccine Alliance (Gavi) introduced PCV into their national immunization programmes. Fifty-seven countries have now introduced PCV since 2010, of which three countries have transitioned from Gavi support as of 2016, and self-finance their vaccine procurement with access to favourable pricing terms established through the Advance Market Commitment (AMC). UNICEF anticipates another one other Gavi approved country to introduce PCV during 2016-2017. • PCV demand from countries eligible to access prices and quantities under the AMC continues to increase. In 2014 and 2015, UNICEF procured a total of 100 and 133 million doses respectively. There are fifteen remaining countries eligible to apply to Gavi for access to financing and/or AMC prices. • The lowest AMC price per dose is currently between US$ 3.30 to US$ 3.50, depending on the product. The price per dose for non-Gavi middle-income countries (MICs) that procure PCV through UNICEF can range from US$ 7.00 to US$ 26.87 per dose. UNICEF will continue to improve the transparency of prices secured for all countries including for MICs. -

COVID-19 Vaccines: Systematic Review
COVID-19 Vaccines: Systematic review Heena Bholaram Choudhary *, Indra Hemaram Sirvi, Yash Rajendra Bamb, Priyanka Rajendra Bamb and Rohan RajkumarPatekar Rasiklal M. Dhariwal Institute of Pharmaceutical Education and Research; Chinchwad, Pune-411019, MH- India. World Journal of Advanced Research and Reviews, 2021, 10(01), 143–155 Publication history: Received on 20 February2021; revised on 01 April 2021; accepted on 03 April 2021 Article DOI: https://doi.org/10.30574/wjarr.2021.10.1.0118 Abstract World facing pandemic challenges of SARS-Co-2 (Severe Acute Respiratory Syndrome Corona Virus 2). Tyrrell and Bynoe identified first strain of human corona virus in 1965. Infection was originated from Wuhan city of China on November 2019. Virus officially named as COVID-19 which causes respiratory tract infections in humans. Preliminary symptoms like chest pain with difficulty in breathing, reduced in systemic oxygen level, cough and fever. At present, the majority of the pharmaceutical companies were in race to developing antiviral drug and vaccines against COVID-19. Industries conducting the experiment and clinical trial of various vaccines to ensure effectiveness and safety on human. Several of them were documentation for patent, authorization of clinical trial and marketing. We provide an overview of Covid-19 history, types of vaccine and mechanism of actions, current future therapeutic drugs and patents. Relative with the mechanism of actions such as inactivated vaccine, vector vaccines, DNA and RNA based vaccine and antiviral drugs already entered in (Phase II and III) clinical trials. The systemic study specified brief summary of various vaccines. Current status of clinical trial, safety and effectiveness according to ICH guidelines. -

Federal Register/Vol. 84, No. 59/Wednesday, March 27, 2019
Federal Register / Vol. 84, No. 59 / Wednesday, March 27, 2019 / Proposed Rules 11473 coating removal to professional users in Environmental Defense Fund. EPA–HQ– effects on children pursuant to the UK, given the requirements for OPPT–2016–0231–0912. Executive Order 13045, entitled limited access that are in place there? 6. Public Comment. DoD Comments on MeCl ‘‘Protection of Children from and NMP 19 Jan 17 Notice of Proposed Environmental Health Risks and Safety IV. Request for Comment and Rulemaking Methylene Chloride and N- Risks’’ (62 FR 19885, April 23, 1997); or Additional Information Methylpyrrolidone; Rulemaking under TSCA Section 6(a). EPA–HQ–OPPT– to consider human health or EPA is seeking comment on all 2016–0231–0519. environmental effects on minority or information outlined in this ANPRM 7. Halogenated Solvents Industry Alliance. low-income populations pursuant to and any other information, which may Responsibly Regulating Methylene Executive Order 12898, entitled not be included in this notice, but Chloride in Paint Removal Products: an ‘‘Federal Actions to Address which you believe is important for EPA Alternative Approach to Flawed Environmental Justice in Minority to consider. Proposal Published by EPA on January Populations and Low-Income EPA specifically invites public 19, 2017. Populations’’ (59 FR 7629, February 16, comment and any additional 8. EPA. How to Get Certified as a Pesticide 1994). Applicator. https://www.epa.gov/ information in response to the questions The Agency will consider such and issues identified in Unit III. pesticide-worker-safety/how-get- certified-pesticide-applicator. Accessed comments during the development of Instructions for providing written December 18, 2018. -

Kaiser Permanente Research Brief Vaccines This Brief Summarizes the Contributions of Kaiser Permanente Research Since 2007 on the Topic of Vaccines
Kaiser Permanente Research Brief Vaccines This brief summarizes the contributions of Kaiser Permanente Research since 2007 on the topic of vaccines. This includes vaccinations delivered in early childhood as well as those delivered to adolescents and adults. Although the development of vaccina- tions against communicable diseases Kaiser Permanente Publications dates back to the 18th century, the Related to Vaccines since 2007 creation of modern vaccines and their widespread use in the United States began in the 20th century.1 Today, the Advisory Committee on Immunization Practices of the Centers for Disease Con- 586 14,000 Journal Articles Citationsa trol and Prevention provides national rec- ommendations for 15 vaccines, including pertussis (whooping cough) shots for pregnant women and numerous vaccina- tions delivered to children, adolescents, 2 and adults (see figure below). Although 123Practice Clinical80 Decision c Guideline Aid References vaccination has largely eliminated dis- b eases such as measles and rubella in the References United States,1 these and others are still found frequently in countries with lower Source: Kaiser Permanente Publications 3 rates of vaccine coverage, and the ease Library and PLUM metrics, as of October 19, of global travel has led to cases in which 2020. a Number of citing journal articles, unvaccinated persons from well-vac- according to Scopus. cinated nations have contracted these b Number of references in PubMed infections.4 Moreover, diseases such as guidelines. c Citations in DynaMed Plus, a point-of-care 1 5 pertussis, varicella (chicken pox) and clinical reference tool. human papillomavirus (HPV)6 still occur frequently in the United States. In addition to the direct effect of immunizing patients against dangerous dis- eases, vaccination also benefits society more broadly through so-called “herd immunity” effects. -

A Dually Active Anthrax Vaccine That Confers Protection Against Both Bacilli and Toxins
A dually active anthrax vaccine that confers protection against both bacilli and toxins Gi-Eun Rhie†‡, Michael H. Roehrl§, Michael Mourez†, R. John Collier†, John J. Mekalanos†, and Julia Y. Wang¶ʈ Departments of †Microbiology and Molecular Genetics and §Biological Chemistry and Molecular Pharmacology, and ¶Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115 Contributed by John J. Mekalanos, July 17, 2003 Systemic anthrax is caused by unimpeded bacillar replication and it consists of D-glutamic acid residues linked by ␥ peptide bonds, toxin secretion. We developed a dually active anthrax vaccine and thus bears no resemblance to mammalian host molecules. (DAAV) that confers simultaneous protection against both bacilli DAAV was designed as a PGA–PA conjugate vaccine to and toxins. DAAV was constructed by conjugating capsular poly- optimize the immunogenicity of both components, especially of ␥-D-glutamic acid (PGA) to protective antigen (PA), converting the the poorly immunogenic PGA (11), because covalent linking of weakly immunogenic PGA to a potent immunogen, and synergis- epitopes often significantly enhances a specific immune response tically enhancing the humoral response to PA. PGA-specific anti- (12). Recombinant PA from Escherichia coli and PGA from bodies bound to encapsulated bacilli and promoted the killing of Bacillus licheniformis ATCC 9945a were prepared, and conju- bacilli by complement. PA-specific antibodies neutralized toxin gates were synthesized by coupling carboxyl groups of PGA to activity and protected immunized mice against lethal challenge the amines of PA by carbodiimide-mediated condensation. We with anthrax toxin. Thus, DAAV combines both antibacterial and present results for two sets of conjugates with 1:2 and 1:1 antitoxic components in a single vaccine against anthrax.