Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

The Oxford – Cambridge Arc Home of the New Innovation Economy
Economic Vision: The Oxford – Cambridge Arc Home of the New Innovation Economy April 2019 Contents 1 Introduction 3 2 The Economic Vision 8 3 The New Innovation Economy: Sectors 11 4 The Innovation & Growth Network 24 5 Achieving Ambitions 29 6 Conclusion: Critical Mass 35 | Introduction 1 Introduction 1.1 This vision’s purpose The purpose of the Economic Vision is to explain the Oxford - Cambridge Arc’s unified proposition as a globally leading innovation and growth catalyst. The Arc offers access to each of the critical ingredients for business and innovation-led growth. This collective offer represents a powerful and coherent expression of the region’s current assets and future potential. This Economic Vision for the Arc sets out an ambition and series of proposals designed to unlock the economic potential of the region and deliver transformative growth for the UK between now and 2050. It provides a vision for how the Arc can better connect its unique and world-leading assets to become truly globally competitive in frontier markets, both for business investment and for top talent. With a bolder brand and stronger international presence the Arc can continue to lead the whole of the UK to the forefront of global innovation excellence in the coming years and decades. 3 | Introduction This Economic Vision is built upon the foundation of This Economic Vision has been developed in the four local industrial strategies which currently partnership with the three LEPs and the Combined demarcate the Arc’s geographic area. These have Authority, who have been given a mandate by Central been prepared by the Oxfordshire (OxLEP), South Government to drive forwards the Economic Vision for East Midlands (SEMLEP) and Buckinghamshire the Arc: Thames Valley (BTVLEP) Local Enterprise Partnerships, as well as the Cambridgeshire & Peterborough Mayoral Combined Authority (CPCA). -

Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Spatiotemporal control of CRISPR/Cas9 gene editing Chenya Zhuo1, Jiabin Zhang1, Jung-Hwan Lee2, Ju Jiao3, Du Cheng4, Li Liu5, Hae-Won Kim2,YuTao1 and Mingqiang Li 1,6 The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/Cas9) gene editing technology, as a revolutionary breakthrough in genetic engineering, offers a promising platform to improve the treatment of various genetic and infectious diseases because of its simple design and powerful ability to edit different loci simultaneously. However, failure to conduct precise gene editing in specific tissues or cells within a certain time may result in undesirable consequences, such as serious off-target effects, representing a critical challenge for the clinical translation of the technology. Recently, some emerging strategies using genetic regulation, chemical and physical strategies to regulate the activity of CRISPR/Cas9 have shown promising results in the improvement of spatiotemporal controllability. Herein, in this review, we first summarize the latest progress of these advanced strategies involving cell-specific promoters, small-molecule activation and inhibition, bioresponsive delivery carriers, and optical/thermal/ultrasonic/magnetic activation. Next, we highlight the advantages and disadvantages of various strategies and discuss their obstacles and limitations in clinical translation. Finally, we propose viewpoints on directions that can be explored to -

Minutes of the Proceedings
MINUTES OF THE PROCEEDINGS at the Thirty‐sixth Meeting of the COUNCIL of the IMPERIAL COLLEGE OF SCIENCE, TECHNOLOGY AND MEDICINE The Thirty‐sixth Meeting of the Council was held in the Council Room, 170 Queen’s Gate, South Kensington Campus, Imperial College London, at 10:00 a.m. on Friday, 16th May 2014, when there were present: The Baroness Manningham‐Buller (Chair), Professor A. Anandalingam, Mr. C. Brinsmead, Dame Ruth Carnall, Mrs. P. Couttie, Professor M.J. Dallman, Mr. P. Dilley, Mr. D. Goldsmith, Professor N. Gooderham, Professor Dame Julia Higgins, Professor D.P.A. Kelleher, Ms. J.R. Lomax, Professor J. Magee, Mr. J. Newsum, Mr. S. Newton, Ms. K. Owen, Mr. M. Sanderson, Professor J Stirling, the President & Rector and the Clerk to the Court and Council. Apologies Mr. I. Conn. MINUTES Council – 7th February 2014 1. The Minutes of the thirty‐fifth Meeting of the Council, held on Friday, 7th February 2014, were taken as read, confirmed and signed. CHAIR’S REPORT 2. The Chair advised members that, following his appointment as Chair of the Chelsea and Westminster Hospital NHS Foundation Trust, Sir Thomas Hughes‐Hallett had resigned as a co‐opted member of the Council on 11 February. PRESIDENT & RECTOR’S REPORT 3. The President & Rector reported that this year’s Imperial Festival, held on 9th and 10th May, had received many visitors at the South Kensington campus to enjoy the various demonstrations, talks 1 Council 16th May 2014 and other activities on offer. What had started as a relatively modest pilot project in 2012 to explore how Imperial College London might share its research with more people had now evolved into a large‐scale and prominent annual fixture in the College’s calendar. -

Medical Research Council Annual Report and Accounts 2006/07 HC 93
06/07 Annual Report and Accounts © Crown Copyright 2006 The text in this document (excluding any Royal Arms and departmental logos) may be reproduced free of charge in any format or medium providing that it is reproduced accurately and not used in a misleading context. The material must be acknowledged as Crown copyright and the title of the document specified. Any queries relating to the copyright in this document should be addressed to The Licensing Division, HMSO, St Clements House, 2-16 Colegate, Norwich, NR3 1BQ. Fax: 01603 723000 or e-mail: licensing@cabinet-office.x.gsi.gov.uk 2 MRC Annual Report and Accounts 2006/07 Medical Research Council Annual Report and Accounts 2006/07 Presented to Parliament by the Secretary of State, and by the Comptroller and Auditor General in pursuance of Schedule I, Sections 2(2) and 3(3) of the Science and Technology Act 1965. Sir John Chisholm Chairman Professor Sir Leszek Borysiewicz Deputy Chairman and Chief Executive Ordered by and printed on London: The Stationery Office 6 February 2008 Price: £18.55 HC 93 The Medical Research Council The MRC RCUK The Medical Research Council (MRC) was set up in 1913 to administer Research Councils UK (RCUK) is a partnership of the seven (formerly public funds for medical research. It was incorporated under its eight) UK Research Councils – public bodies funded mainly by the UK present title by Royal Charter in 1920. A supplemental charter was Government via OSI. granted in 1993 describing the MRC’s new mission following the 1993 government white paper on science and technology. -
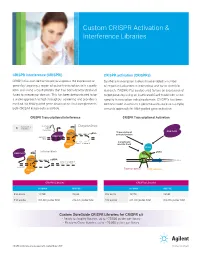
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

Michael John Owen Wakelam 1955–2020
obituary Michael John Owen Wakelam 1955–2020 We have lost a distinguished biochemist who dedicated his career to the study of phosphatidylinositol signalling in metabolic regulation and to the advancement of lipidomics. ichael John Owen Wakelam passed away on 31 March, 2020 at the Mage of 64, much too early. He became known to colleagues and friends as a scientist highly regarded for his research. He was honoured in 2018 with the Morton Lectureship of the Biochemical Society and was elected a fellow of the Royal Society of Biology. In 2019, he was elected a member of the Academia Europaea. Academic career It was not by chance that Michael turned his focus to phosphatidylinositols (PIs) and lipid signalling. He was a student and later a professor at the University of Birmingham at a time when Britain was the world’s hub of pioneering phospholipid research, such as that by J. N. Hawthorne and Bob Michell on PI in Birmingham. Bob Michell inspired Michael extraordinarily, and they even published a joint review. Subsequently, Michael moved to Cambridge to serve as the director of the Babraham Institute, where Credit: Babraham Institute Rex Dawson and Robin Irvine studied phospholipids, particularly PI metabolism and signalling. A joint publication by Michael and Irvine appeared as well. the Babraham Institute, and he received an the Keystone Symposia on Molecular and Certainly, Michael’s scientific engagement honorary professorship in lipid signalling at Cellular Biology. can be seen in the tradition of these eminent the University of Cambridge Clinical School. phospholipid scientists. In addition, he was an honorary professor Lipid signalling and metabolism Michael’s career path is impressive. -

A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
METHODS, MODELS & TECHNIQUES Prospects & Overviews www.bioessays-journal.com A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities Yang Liu and Baojun Wang* friend or foe” system, scientists are CRISPR (clustered regularly interspaced short palindromic repeats) activation able to guide the endonucleases to (CRISPRa) in bacteria is an attractive method for programmable gene their desired DNA or RNA targets.[5–8] activation. Recently, a eukaryote-like, 54-dependent CRISPRa system has CRISPR regulation relies on inactivated been reported. It exhibits high dynamic ranges and permits flexible target site CRISPR endonucleases. The nuclease- deficient CRISPR DNA endonucleases, selection. Here, an overview of the existing strategies of CRISPRa in bacteria for instances dCas9 and ddCpf1 (dCas12), is presented, and the characteristics and design principles of the CRISPRa are effectively programmable DNA bind- system are introduced. Possible scenarios for applying the eukaryote-like ing domains. When these domains are CRISPRa system is discussed with corresponding suggestions for tethered to transactivation domains or performance optimization and future functional expansion. The authors subunits of RNA polymerase, they ac- envision the new eukaryote-like CRISPRa system enabling novel designs in tivate the promoters near the CRISPR target sites. This strategy has been 54 multiplexed gene regulation and promoting research in the -dependent widely utilized in both eukaryotes and gene regulatory networks among a variety of biotechnology relevant or prokaryotes,[9–17] particularly in the former, disease-associated bacterial species. where the transcription activation mecha- nisms and the activators are well-studied. While CRISPRa in eukaryotes enjoys much success and is continuously im- 1. -

24, 2021 Program Book Table of Contents
June 21 – 24, 2021 Program Book Table of Contents Genetics Society of America . 3 Conference Organizers . 5 International C. elegans Board 2021 . 7 Sponsors . 9 Schedule of Events . 11 General Information . 16 Conference App . .. 17 Oral Presenters . 17 Poster Presenters . 17 Viewing Oral Sessions . 18 Attending Live Poster Sessions . .. 18 Live Poster Session Schedule . 19 Sponsor and Exhibitor Education Sessions . 21 Daily Meet-ups via Zoom and Remo . 22 Viewing Virtual Posters on the App . 23 Slack Chat Channels . 23 Job Postings . 23 Presenting Author Index . 23 Conference Policies . 24 Exhibits . 27 Oral Presentation and Workshop Session Listings . 29 Poster Session Listings . 58 23rd International C. elegans Conference | 2 Genetics Society of America Genetics Society of America GSA is an international scientific society representing more than 5,000 researchers and educators around the world. As well as connecting researchers through conferences and career programs, we publish two peer- edited scholarly journals, GENETICS and G3: Genes|Genomes|Genetics. We encourage you to join GSA so you can make use of exclusive member benefits and get involved in the Society’s many programs, including professional development training, awards, advocacy, and more. Join us as we work to advance the field and serve our community. Visit genetics-gsa.org for more information. GENETICS has been innovating since 1916, publishing high quality original research across the breadth of the field. G3: Genes|Genomes|Genetics is an open access journal that publishes high quality, useful results regardless of perceived impact. 2021 GSA Board of Directors Officers Directors Journal Editors Hugo Bellen, President Swathi Arur Brenda J. -

Crp-Brochure-080518-Na-Web.Pdf
Where Life Meets Science Where Life Meets Science “Research excellence is one of the The Park offers a flexible and fostering CHESTERFORD RESEARCH PARK Building on 60 years of continuous R&D at Chesterford is being developed as a 250 Foreword defining features of the Cambridge environment for both established R&D PROVIDES A SUPERBLY FLEXIBLE Chesterford, innovative biotechnology and acre low density scheme. To date, more landscape. Both academically and in companies and start-ups alike, providing AND FUTURE PROOFED pharmaceutical occupiers thrive in than 300,000 sq ft of laboratory and R&D the transfer of knowledge through to cutting-edge research facilities within a state-of-the-art accommodation and enjoy space has been let and occupied. Further ENVIRONMENT FOR BOTH EARLY commercial application, our region has community that encourages collaboration modern, central facilities, all set within a phases of construction are proposed to STAGE AND ESTABLISHED R&D established an international reputation at every stage. Chesterford Research Park unique and idyllic parkland location. extend the development to approximately that attracts outstanding academics, will help ensure that Cambridge ideas COMPANIES 1 million sq ft. researchers and business leaders who continue to change the world.” collectively drive discovery in so many different spheres. The depth and amazing Professor Sir Leszek Borysiewicz diversity of that research capability is Vice-Chancellor of the University of Cambridge. apparent at Chesterford Research Park. (Retired 2017) -

Trends and Challenges in Computational RNA Biology Alina Selega and Guido Sanguinetti*
Selega and Sanguinetti Genome Biology (2016) 17:253 DOI 10.1186/s13059-016-1117-7 MEETINGREPORT Open Access Trends and challenges in computational RNA biology Alina Selega and Guido Sanguinetti* Abstract complemented by two lively poster sessions, where partic- ipants had an opportunity to engage with over 40 posters A report on the Wellcome Trust Conference on during evening drinks receptions. Computational RNA Biology, held in Hinxton, UK, on In this report, we briefly recount the content of the 17–19 October 2016. conference by providing condensed, headline-style sum- Keywords: RNA, Review, Computational biology maries of the research described in the talks and some posters. Within the scope of this brief report, we cannot possibly do justice to the wealth and breadth of material Introduction presented and we will not be able to mention much inter- Recent years have witnessed a profound shift in our esting research, particularly within the poster sessions. understanding of RNA biology. Several novel biochemical We would like to stress that omissions in this report are and sequencing techniques are producing vast amounts not based on quality, but simply on a personal judgement of data that fundamentally challenge the textbook view as to what material could be most coherently presented in of RNA as a simple intermediate step of gene expression, a very limited space. revealing a wealth of unexpected new roles and shed- ding light on the complexity of the RNA world. While Transcripts the emerging picture unequivocally points to the cen- Perhaps the most remarkable discovery in modern RNA trality of RNA as a mediator of most cellular functions, biology is the realization of the diversity of the transcrip- the richness and heterogeneity of modern datasets pose tome.