Synthetic CRISPR-Cas Gene Activators for Transcriptional Reprogramming in Bacteria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas, Melanie A. Eckersley-Maslin, Wolf Reik celia.x.aldacatalinas@gsk. com (C.A.-C.) [email protected]. uk (W.R.) Highlights Protocol for CRISPRa screens with single- cell readout to interrogate gene function Detailed description of CRISPRa screening procedures in mouse embryonic stem cells Detailed steps on how to construct derived single-cell sgRNA amplicon libraries CRISPR/Cas9 screens are a powerful approach to identify key regulators of biological processes. By combining pooled CRISPR/Cas9 screening with a single-cell RNA-sequencing readout, individual perturbations can be assessed in parallel both comprehensively and at scale. Importantly, this allows gene function and regulation to be interrogated at a cellular level in an unbiased manner. Here, we present a protocol to perform pooled CRISPR-activation screens in mouse embryonic stem cells using 103 Genomics scRNA-seq as a readout. Alda-Catalinas et al., STAR Protocols 2, 100426 June 18, 2021 ª 2021 The Authors. https://doi.org/10.1016/ j.xpro.2021.100426 ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas,1,4,7,* Melanie A. Eckersley-Maslin,1,5,6 and Wolf Reik1,2,3,8,* 1Epigenetics Programme, Babraham Institute, Cambridge CB22 3AT, UK 2Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK 3Centre for Trophoblast Research, University of -

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

Escherichia Coli: Protein Families and Binding Sites M
Functional determinants of transcription factors in Escherichia coli: protein families and binding sites M. Madan Babu and Sarah A. Teichmann DNA-binding transcription factors regulate the expression A reasonable hypothesis would be to assume that of genes near to where they bind. These factors can be activators, repressors and dual regulators are more activators or repressors of transcription, or both. Thus, a closely related to the other proteins within each fundamental question is what determines whether a regulatory group than to proteins in other groups. This transcription factor acts as an activator or repressor? would mean that there would be protein families and Previous research into this question found that a protein's domain architectures that were characteristic either of regulatory function is determined by one or more of the activators, repressors or dual regulators. Prag et al. [10] following factors: protein–proteinprotein–protein contacts, position of the and Perez-Rueda et al. [11] reported evidence to support DNA-binding domain in the protein primary sequence, this by relating the position of helix-turn-helix motifs altered DNA structure, and the position of its binding site along the primary sequence to a protein's regulatory on the DNA relative to the transcription start site. Although function. They used sequence comparison and profile there are many aspects specific to different transcription methods to locate the helix-turn-helix motifs that factors, in this work we demonstrate that, in general, in the represent DBDs, and found that the helix-turn-helix prokaryote Escherichia coli, a transcription factor's protein tends to be at the N terminus for repressors and at the C family is not indicative of its regulatory function, but the terminus for activators. -

Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Spatiotemporal control of CRISPR/Cas9 gene editing Chenya Zhuo1, Jiabin Zhang1, Jung-Hwan Lee2, Ju Jiao3, Du Cheng4, Li Liu5, Hae-Won Kim2,YuTao1 and Mingqiang Li 1,6 The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/Cas9) gene editing technology, as a revolutionary breakthrough in genetic engineering, offers a promising platform to improve the treatment of various genetic and infectious diseases because of its simple design and powerful ability to edit different loci simultaneously. However, failure to conduct precise gene editing in specific tissues or cells within a certain time may result in undesirable consequences, such as serious off-target effects, representing a critical challenge for the clinical translation of the technology. Recently, some emerging strategies using genetic regulation, chemical and physical strategies to regulate the activity of CRISPR/Cas9 have shown promising results in the improvement of spatiotemporal controllability. Herein, in this review, we first summarize the latest progress of these advanced strategies involving cell-specific promoters, small-molecule activation and inhibition, bioresponsive delivery carriers, and optical/thermal/ultrasonic/magnetic activation. Next, we highlight the advantages and disadvantages of various strategies and discuss their obstacles and limitations in clinical translation. Finally, we propose viewpoints on directions that can be explored to -

Regulondb (Version 6.0): Gene Regulation Model Of
D120–D124 Nucleic Acids Research, 2008, Vol. 36, Database issue doi:10.1093/nar/gkm994 RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation Socorro Gama-Castro1, Vero´ nica Jime´ nez-Jacinto1, Martı´n Peralta-Gil1, Alberto Santos-Zavaleta1,Mo´ nica I. Pen˜ aloza-Spinola1, Bruno Contreras-Moreira1, Juan Segura-Salazar1, Luis Mun˜ iz-Rascado1, Irma Martı´nez-Flores1, Heladia Salgado1,Ce´ sar Bonavides-Martı´nez1, Cei Abreu-Goodger1, Carlos Rodrı´guez-Penagos1, Juan Miranda-Rı´os2, Enrique Morett2, Enrique Merino2, Araceli M. Huerta1, Luis Trevin˜ o-Quintanilla1 and Julio Collado-Vides1,* Downloaded from 1Program of Computational Genomics, Centro de Ciencias Geno´ micas, Universidad Nacional Auto´ noma de Me´ xico, A.P. 565-A, Cuernavaca, Morelos 62100, Mexico and 2Instituto de Biotecnologı´a, Universidad Nacional Auto´ noma de Me´ xico, A.P. 510-3, Cuernavaca, Morelos 62100, Mexico http://nar.oxfordjournals.org/ Received September 14, 2007; Revised October 19, 2007; Accepted October 22, 2007 ABSTRACT with more than 4000 curation notes, can now be RegulonDB (http://regulondb.ccg.unam.mx/) is the searched with the Textpresso text mining engine. primary reference database offering curated knowl- edge of the transcriptional regulatory network at Carleton University on June 21, 2015 of Escherichia coli K12, currently the best-known INTRODUCTION electronically encoded database of the genetic A major current task for bioinformatics is that of repre- regulatory network of any free-living organism. senting biological information into an electronic and This paper summarizes the improvements, new computable form. These computational representations biology and new features available in version 6.0. -
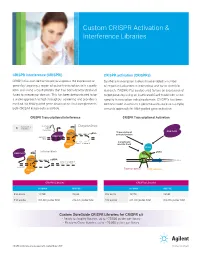
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
METHODS, MODELS & TECHNIQUES Prospects & Overviews www.bioessays-journal.com A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities Yang Liu and Baojun Wang* friend or foe” system, scientists are CRISPR (clustered regularly interspaced short palindromic repeats) activation able to guide the endonucleases to (CRISPRa) in bacteria is an attractive method for programmable gene their desired DNA or RNA targets.[5–8] activation. Recently, a eukaryote-like, 54-dependent CRISPRa system has CRISPR regulation relies on inactivated been reported. It exhibits high dynamic ranges and permits flexible target site CRISPR endonucleases. The nuclease- deficient CRISPR DNA endonucleases, selection. Here, an overview of the existing strategies of CRISPRa in bacteria for instances dCas9 and ddCpf1 (dCas12), is presented, and the characteristics and design principles of the CRISPRa are effectively programmable DNA bind- system are introduced. Possible scenarios for applying the eukaryote-like ing domains. When these domains are CRISPRa system is discussed with corresponding suggestions for tethered to transactivation domains or performance optimization and future functional expansion. The authors subunits of RNA polymerase, they ac- envision the new eukaryote-like CRISPRa system enabling novel designs in tivate the promoters near the CRISPR target sites. This strategy has been 54 multiplexed gene regulation and promoting research in the -dependent widely utilized in both eukaryotes and gene regulatory networks among a variety of biotechnology relevant or prokaryotes,[9–17] particularly in the former, disease-associated bacterial species. where the transcription activation mecha- nisms and the activators are well-studied. While CRISPRa in eukaryotes enjoys much success and is continuously im- 1. -

Description Data File Programa De Genómica Computacional / Centro De Ciencias Genómicas
Description Data File Programa de Genómica Computacional / Centro de Ciencias Genómicas DESCRIPTION DATA FILE 1. GENERAL INFORMATION. Title: LeuO and H-NS Data Set Reference: Shimada T, Bridier A, Briandet R, Ishihama A. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol. 2011 Oct;82(2):378-97. PMID: 21883529 Contact person for this data set: Questions concerning the content of the data set that are raised by users of RegulonDB will be forwarded to this person. We would appreciate receiving a copy of the response to the user, so we can keep track of taking care of user requests. Person: RegulonDB staff Email address: [email protected] 2. DATA SET DESCRIPTION. Summary: LeuO Data Set. LeuO, the regulator of the leucine biosynthesis operon of Escherichia coli, is involved in the regulation of as-yet-unspecified genes that affect the stress response and pathogenesis expression. LeuO is involved in an antagonistic interplay with the universal silencer H-NS. Genomic SELEX screening was performed to identify the whole set of LeuO and H-NS regulation targets. A total of 140 LeuO-binding peaks (183 regulatory interactions) were identified by SELEX methodology, of which as many as 133 (95%) were found to contain the binding site of H-NS. In addition, a DNA microarray assay was carried out to identify genes affected by deletion or overexpression of the leuO gene. A total of 35 regulatory interactions were found via SELEX as well as microarray analysis. Based on the behavior of the regulated genes in the microarray analysis, from which it was determined that LeuO acts as an activator for 18 of them, 13 as a repressor and 4 as a dual function; we deduced that LeuO acts as a dual regulator. -

Protein-DNA Interactions II
Protein-DNA Interactions II Bio 5488 Overview 1. Review of information content and weight matrices 2. Online PWM resources 3. Motif discovery: Greedy algorithm 4. Motif discovery: Gibbs sampling Information Content EcoR1 Random Rap1 GAATTC GCCTAC TGTATGGGTG GAATTC ACATTC TGTTCGGATT GAATTC TCATTC TGCATGGGTG GAATTC CGACTC TGTACAGGTG GAATTC GAATTC TGTATGGATG GAATTC ATATCG TGTTCGGGTT GAATTC GAAATG TGTATGGGTG Information Content Matrix of Frequencies A: 0.1 0.7 0.2 0.3 0.4 0.1 C: 0.1 0.1 0.1 0.3 0.2 0.1 G: 0.1 0.1 0.2 0.1 0.2 0.1 T: 0.7 0.1 0.5 0.3 0.2 0.7 aka Relative Entropy, Kullbach-Liebler Distance Obtaining a Weight Matrix Hertz and Stormo, Bioinformatics (1999) Online tools: PWMs JASPAR (jaspar.genereg.net) open-access curated, non-redundant, PWMs for multi-cellular eukaryotes hundreds of PWMs JASPAR_FAM - metamodels for structural families Online tools: PWMs AC M00213 XX ID F$RAP1_C XX DT 05.09.1995 (created); dbo. DT 30.11.1995 (updated); ewi. CO Copyright (C), Biobase GmbH. XX NA RAP1 XX DE yeast repressor/activator protein 1 XX BF T00715 RAP1; Species: yeast, Saccharomyces cerevisiae. XX PO A C G T 01 6.89 2.40 0.79 4.74 W 02 8.80 0.00 4.58 1.43 R 03 7.20 6.83 0.79 0.00 M 04 14.82 0.00 0.00 0.00 A TRANSFAC 05 0.00 14.82 0.00 0.00 C 06 0.00 14.82 0.00 0.00 C Free access w/ reduced functionality 07 0.00 14.82 0.00 0.00 C 08 14.82 0.00 0.00 0.00 A to professional version 09 2.13 0.67 2.87 9.15 T 10 11.92 0.76 1.49 0.64 A 11 0.76 14.06 0.00 0.00 C 12 11.45 3.36 0.00 0.00 A Public version dates to 2005 13 0.79 5.64 0.00 7.10 Y 14 1.64 7.18 1.61 4.39 Y XX BA total weight of sequences: 14.82 XX CC consind generated matrix (random_expectation: 0.03) XX // (www.gene-regulation.com/pub/databases.html/#transfac) Online tools: PWMs RegulonDB (regulondb.ccg.unam.mx) Bacterial regulons and operons in E. -

A Database of Regulatory Networks in Gamma-Proteobacterial Genomes Abel D
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Open Repository and Bibliography - Luxembourg D98–D102 Nucleic Acids Research, 2005, Vol. 33, Database issue doi:10.1093/nar/gki054 TRACTOR_DB: a database of regulatory networks in gamma-proteobacterial genomes Abel D. Gonza´lez, Vladimir Espinosa, Ana T. Vasconcelos1, Ernesto Pe´rez-Rueda2 and Julio Collado-Vides3,* National Bioinformatics Center, Industria y San Jose´, Capitolio Nacional, CP. 10200, Habana Vieja, Habana, Cuba, 1National Laboratory for Scientific Computing, Avenue Getulio Vargas 333, Quitandinha, CEP 25651-075, Petropolis, Rio de Janeiro, Brazil, 2Depto. de Ingenierı`a Celular y Biocata´lisis, IBT-UNAM, Cuernavaca, Morelos, Mexico and 3Center of Genomics, UNAM, AP 565-A Cuernavaca, CP. 62100, Morelos, Mexico Received August 6, 2004; Revised and Accepted October 1, 2004 Downloaded from ABSTRACT out in this direction (1–3). The first step towards this goal is the recognition of all the genes regulated by a transcription factor Experimental data on the Escherichia coli (TF), i.e. its regulon. transcriptional regulatory system has been used in Computational approaches to recognizing the location of http://nar.oxfordjournals.org/ the past years to predict new regulatory elements regulatory sites in bacterial genomes include the use of weight (promoters, transcription factors (TFs), TFs’ binding matrices (4), phylogenetic footprinting (5), searching for sites and operons) within its genome. As more gen- statistical overrepresentation of oligonucleotides within a omes of gamma-proteobacteria are being sequenced, genome and clustering co-expressed genes in order to find the prediction of these elements in a growing number conserved patterns in their upstream regions (6,7), among of organisms has become more feasible, as a others (8,9). -

A User-Friendly Tool for Improving Bacterial Genome Annotation Through Analysis of Transcription Control Signals
SigmoID: a user-friendly tool for improving bacterial genome annotation through analysis of transcription control signals Yevgeny Nikolaichik and Aliaksandr U. Damienikan Department of Molecular Biology, Belarusian State University, Minsk, Belarus ABSTRACT The majority of bacterial genome annotations are currently automated and based on a `gene by gene' approach. Regulatory signals and operon structures are rarely taken into account which often results in incomplete and even incorrect gene function assignments. Here we present SigmoID, a cross-platform (OS X, Linux and Windows) open-source application aiming at simplifying the identification of transcription regulatory sites (promoters, transcription factor binding sites and terminators) in bac- terial genomes and providing assistance in correcting annotations in accordance with regulatory information. SigmoID combines a user-friendly graphical interface to well known command line tools with a genome browser for visualising regulatory elements in genomic context. Integrated access to online databases with regulatory information (RegPrecise and RegulonDB) and web-based search engines speeds up genome analysis and simplifies correction of genome annotation. We demonstrate some features of SigmoID by constructing a series of regulatory protein binding site profiles for two groups of bacteria: Soft Rot Enterobacteriaceae (Pectobacterium and Dickeya spp.) and Pseudomonas spp. Furthermore, we inferred over 900 transcription factor binding sites and alternative sigma factor promoters in the annotated genome of Pectobacterium atrosepticum. These regulatory signals control putative transcription units covering about 40% of the P. atrosepticum chromosome. Reviewing the annotation in cases where Submitted 26 January 2016 it didn't fit with regulatory information allowed us to correct product and gene names Accepted 29 April 2016 for over 300 loci.