Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas, Melanie A. Eckersley-Maslin, Wolf Reik celia.x.aldacatalinas@gsk. com (C.A.-C.) [email protected]. uk (W.R.) Highlights Protocol for CRISPRa screens with single- cell readout to interrogate gene function Detailed description of CRISPRa screening procedures in mouse embryonic stem cells Detailed steps on how to construct derived single-cell sgRNA amplicon libraries CRISPR/Cas9 screens are a powerful approach to identify key regulators of biological processes. By combining pooled CRISPR/Cas9 screening with a single-cell RNA-sequencing readout, individual perturbations can be assessed in parallel both comprehensively and at scale. Importantly, this allows gene function and regulation to be interrogated at a cellular level in an unbiased manner. Here, we present a protocol to perform pooled CRISPR-activation screens in mouse embryonic stem cells using 103 Genomics scRNA-seq as a readout. Alda-Catalinas et al., STAR Protocols 2, 100426 June 18, 2021 ª 2021 The Authors. https://doi.org/10.1016/ j.xpro.2021.100426 ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas,1,4,7,* Melanie A. Eckersley-Maslin,1,5,6 and Wolf Reik1,2,3,8,* 1Epigenetics Programme, Babraham Institute, Cambridge CB22 3AT, UK 2Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK 3Centre for Trophoblast Research, University of -

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

Editing DNA Methylation in Mammalian Embryos
International Journal of Molecular Sciences Review Editing DNA Methylation in Mammalian Embryos Taiga Yamazaki 1,* , Yu Hatano 2, Ryoya Taniguchi 2, Noritada Kobayashi 1 and Kazuo Yamagata 2,* 1 Division of Biomedical Research, Kitasato University Medical Center, Kitasato University, 6-100 Arai, Kitamoto, Saitama 364-8501, Japan; [email protected] 2 Faculty of Biology-Oriented Science and Technology, KINDAI University, 930 Nishimitani, Kinokawa, Wakayama 649-6493, Japan; [email protected] (Y.H.); [email protected] (R.T.) * Correspondence: [email protected] (T.Y.); [email protected] (K.Y.); Tel.:+81-48-593-1212 (T.Y.), +81-736-77-3888 (K.Y.) Received: 11 December 2019; Accepted: 16 January 2020; Published: 18 January 2020 Abstract: DNA methylation in mammals is essential for numerous biological functions, such as ensuring chromosomal stability, genomic imprinting, and X-chromosome inactivation through transcriptional regulation. Gene knockout of DNA methyltransferases and demethylation enzymes has made significant contributions to analyzing the functions of DNA methylation in development. By applying epigenome editing, it is now possible to manipulate DNA methylation in specific genomic regions and to understand the functions of these modifications. In this review, we first describe recent DNA methylation editing technology. We then focused on changes in DNA methylation status during mammalian gametogenesis and preimplantation development, and have discussed the implications of applying this technology to early embryos. Keywords: DNA methylation; epigenome editing; preimplantation embryo; germ cell; centromere; pericentromere 1. Introduction Cytosine methylation is a process in which methyl groups are added to the cytosine of CpG dinucleotides, forming 5-methylcytosine (5mC). -

Strategies for Precision Modulation of Gene Expression by Epigenome Editing: an Overview Benjamin I
Laufer and Singh Epigenetics & Chromatin (2015) 8:34 DOI 10.1186/s13072-015-0023-7 REVIEW Open Access Strategies for precision modulation of gene expression by epigenome editing: an overview Benjamin I. Laufer* and Shiva M. Singh Abstract Genome editing technology has evolved rather quickly and become accessible to most researchers. It has resulted in far reaching implications and a number of novel designer systems including epigenome editing. Epigenome editing utilizes a combination of nuclease-null genome editing systems and effector domains to modulate gene expression. In particular, Zinc Finger, Transcription-Activator-Like Effector, and CRISPR/Cas9 have emerged as modular systems that can be modified to allow for precision manipulation of epigenetic marks without altering underlying DNA sequence. This review contains a comprehensive catalog of effector domains that can be used with components of genome editing systems to achieve epigenome editing. Ultimately, the evidence-based design of epigenome editing offers a novel improvement to the limited attenuation strategies. There is much potential for editing and/or correcting gene expression in somatic cells toward a new era of functional genomics and personalized medicine. Keywords: Regulation of gene expression, Functional genomics, Stem cells, dCas9, CRISPR/Cas9, Zinc Finger, Transcription-Activator-Like Effector (TALE), Synthetic biology Background editing systems rely on two components, a DNA-bind- The modulation of gene expression can be achieved by ing element, and nuclease, to modify the targeted DNA a variety of biotechnologies such as RNA interference, sequence. Genome editing can be used to study protein non-precision drugs, and artificial transcription factors function by altering coding sequence or achieve tran- (ATFs). -
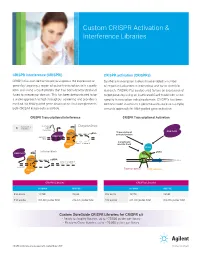
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
METHODS, MODELS & TECHNIQUES Prospects & Overviews www.bioessays-journal.com A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities Yang Liu and Baojun Wang* friend or foe” system, scientists are CRISPR (clustered regularly interspaced short palindromic repeats) activation able to guide the endonucleases to (CRISPRa) in bacteria is an attractive method for programmable gene their desired DNA or RNA targets.[5–8] activation. Recently, a eukaryote-like, 54-dependent CRISPRa system has CRISPR regulation relies on inactivated been reported. It exhibits high dynamic ranges and permits flexible target site CRISPR endonucleases. The nuclease- deficient CRISPR DNA endonucleases, selection. Here, an overview of the existing strategies of CRISPRa in bacteria for instances dCas9 and ddCpf1 (dCas12), is presented, and the characteristics and design principles of the CRISPRa are effectively programmable DNA bind- system are introduced. Possible scenarios for applying the eukaryote-like ing domains. When these domains are CRISPRa system is discussed with corresponding suggestions for tethered to transactivation domains or performance optimization and future functional expansion. The authors subunits of RNA polymerase, they ac- envision the new eukaryote-like CRISPRa system enabling novel designs in tivate the promoters near the CRISPR target sites. This strategy has been 54 multiplexed gene regulation and promoting research in the -dependent widely utilized in both eukaryotes and gene regulatory networks among a variety of biotechnology relevant or prokaryotes,[9–17] particularly in the former, disease-associated bacterial species. where the transcription activation mecha- nisms and the activators are well-studied. While CRISPRa in eukaryotes enjoys much success and is continuously im- 1. -

Stem Cells, Genome Editing and the Path to Translational Medicine
Stem Cells, Genome Editing, and the Path to Translational Medicine The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Soldner, Frank and Rudolf Jaenisch. "Stem Cells, Genome Editing, and the Path to Translational Medicine." Cell 175, 3 (October 2018): P615-632 © 2018 Elsevier Inc As Published http://dx.doi.org/10.1016/j.cell.2018.09.010 Publisher Elsevier BV Version Author's final manuscript Citable link https://hdl.handle.net/1721.1/125973 Terms of Use Creative Commons Attribution-NonCommercial-NoDerivs License Detailed Terms http://creativecommons.org/licenses/by-nc-nd/4.0/ HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Cell. Author Manuscript Author manuscript; Manuscript Author available in PMC 2019 October 18. Published in final edited form as: Cell. 2018 October 18; 175(3): 615–632. doi:10.1016/j.cell.2018.09.010. Stem cells, genome editing and the path to translational medicine Frank Soldner1 and Rudolf Jaenisch1,2,3 1The Whitehead Institute, 455 Main Street, Cambridge, MA 02142, USA 2Department of Biology, Massachusetts Institute of Technology, 31 Ames Street, Cambridge, MA 02139, USA Summary The derivation of human embryonic stem cells (hESCs) and the stunning discovery that somatic cells can be reprogrammed into human induced pluripotent stem cells (hiPSCs) holds the promise to revolutionize biomedical research and regenerative medicine. In this review, we focus on disorders of the central nervous system and explore how advances in human pluripotent stem cells (hPSCs) coincide with evolutions in genome engineering and genomic technologies to provide realistic opportunities to tackle some of the most devastating complex disorders. -

Challenges, New Technologies and Their Use in Plants
International Journal of Molecular Sciences Review Chromatin Manipulation and Editing: Challenges, New Technologies and Their Use in Plants Kateryna Fal 1,† , Denisa Tomkova 2,†, Gilles Vachon 1 , Marie-Edith Chabouté 2, Alexandre Berr 2,* and Cristel C. Carles 1,* 1 Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS, CEA, INRAE, IRIG-LPCV, 38000 Grenoble, France; [email protected] (K.F.); [email protected] (G.V.) 2 Institut de Biologie Moléculaire des Plantes du CNRS, Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg CEDEX, France; [email protected] (D.T.); [email protected] (M.-E.C.) * Correspondence: [email protected] (A.B.); [email protected] (C.C.C.) † These authors contributed equally to this work. Abstract: An ongoing challenge in functional epigenomics is to develop tools for precise manipulation of epigenetic marks. These tools would allow moving from correlation-based to causal-based findings, a necessary step to reach conclusions on mechanistic principles. In this review, we describe and discuss the advantages and limits of tools and technologies developed to impact epigenetic marks, and which could be employed to study their direct effect on nuclear and chromatin structure, on transcription, and their further genuine role in plant cell fate and development. On one hand, epigenome-wide approaches include drug inhibitors for chromatin modifiers or readers, nanobodies against histone marks or lines expressing modified histones or mutant chromatin effectors. On the other hand, locus-specific approaches consist in targeting precise regions on the chromatin, with engineered proteins able to modify epigenetic marks. -

Concurrent Genome and Epigenome Editing by CRISPR-Mediated Sequence Replacement Jes Alexander1,2, Gregory M
Alexander et al. BMC Biology (2019) 17:90 https://doi.org/10.1186/s12915-019-0711-z METHODOLOGY ARTICLE Open Access Concurrent genome and epigenome editing by CRISPR-mediated sequence replacement Jes Alexander1,2, Gregory M. Findlay1, Martin Kircher1 and Jay Shendure1,3,4,5* Abstract Background: Recent advances in genome editing have facilitated the direct manipulation of not only the genome, but also the epigenome. Genome editing is typically performed by introducing a single CRISPR/Cas9-mediated double- strand break (DSB), followed by non-homologous end joining (NHEJ)- or homology-directed repair-mediated repair. Epigenome editing, and in particular methylation of CpG dinucleotides, can be performed using catalytically inactive Cas9 (dCas9) fused to a methyltransferase domain. However, for investigations of the role of methylation in gene silencing, studies based on dCas9-methyltransferase have limited resolution and are potentially confounded by the effects of binding of the fusion protein. As an alternative strategy for epigenome editing, we tested CRISPR/Cas9 dual cutting of the genome in the presence of in vitro methylated exogenous DNA, with the aim of driving replacement of the DNA sequence intervening the dual cuts via NHEJ. Results: In a proof of concept at the HPRT1 promoter, successful replacement events with heavily methylated alleles of a CpG island resulted in functional silencing of the HPRT1 gene. Although still limited in efficiency, our study demonstrates concurrent epigenome and genome editing in a single event. Conclusions: This study opens the door to investigations of the functional consequences of methylation patterns at single CpG dinucleotide resolution. Our results furthermore support the conclusion that promoter methylation is sufficient to functionally silence gene expression. -

Novel Epigenetic Techniques Provided by the CRISPR/Cas9
Hindawi Stem Cells International Volume 2018, Article ID 7834175, 12 pages https://doi.org/10.1155/2018/7834175 Review Article Novel Epigenetic Techniques Provided by the CRISPR/ Cas9 System 1,2 1,2 1,2 1,3,2 Nina Xie , Yafang Zhou, Qiying Sun , and Beisha Tang 1Department of Geriatric Neurology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China 2National Clinical Research Center for Geriatric Disorders, Central South University, Changsha, Hunan 410078, China 3Department of Neurology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China Correspondence should be addressed to Beisha Tang; [email protected] Received 4 November 2017; Revised 4 February 2018; Accepted 27 March 2018; Published 8 July 2018 Academic Editor: Changwon Park Copyright © 2018 Nina Xie et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Epigenetics classically refers to the inheritable changes of hereditary information without perturbing DNA sequences. Understanding mechanisms of how epigenetic factors contribute to inheritable phenotype changes and cell identity will pave the way for us to understand diverse biological processes. In recent years, the emergence of CRISPR/Cas9 technology has provided us with new routes to the epigenetic field. In this review, novel epigenetic techniques utilizing the CRISPR/Cas9 system are the main contents to be discussed, including epigenome editing, temporal and spatial control of epigenetic effectors, noncoding RNA manipulation, chromatin in vivo imaging, and epigenetic element screening. 1. Introduction RNA, researchers could target Cas9 nuclease to almost any locus in the genome precisely. -

Chemical and Light Inducible Epigenome Editing
International Journal of Molecular Sciences Review Chemical and Light Inducible Epigenome Editing Weiye Zhao , Yufan Wang and Fu-Sen Liang * Department of Chemistry, Case Western Reserve University 2080 Adelbert Road, Cleveland, OH 44106, USA; [email protected] (W.Z.); [email protected] (Y.W.) * Correspondence: [email protected] Received: 22 December 2019; Accepted: 30 January 2020; Published: 3 February 2020 Abstract: The epigenome defines the unique gene expression patterns and resulting cellular behaviors in different cell types. Epigenome dysregulation has been directly linked to various human diseases. Epigenome editing enabling genome locus-specific targeting of epigenome modifiers to directly alter specific local epigenome modifications offers a revolutionary tool for mechanistic studies in epigenome regulation as well as the development of novel epigenome therapies. Inducible and reversible epigenome editing provides unique temporal control critical for understanding the dynamics and kinetics of epigenome regulation. This review summarizes the progress in the development of spatiotemporal-specific tools using small molecules or light as inducers to achieve the conditional control of epigenome editing and their applications in epigenetic research. Keywords: epigenome; epigenome modifications; chemically induced proximity; light control 1. Introduction Chemical modification patterns on histone tails and DNA collectively constitute the epigenome that dictates unique gene expression patterns and resulting phenotypes in each distinct cell type [1–3]. The alterations of the epigenome due to the dysregulation of epigenome pathways and mutations of epigenome regulators contributes to pathogenesis of various human diseases including many developmental diseases and cancers [4–19]. Several epigenetic regulatory mechanisms including DNA methylations, histone tail posttranslational modifications (PTMs), ATP-dependent chromatin remodeling and non-coding RNAs have been shown to play key roles in governing gene activities [6,7,15,20–29].