A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas, Melanie A. Eckersley-Maslin, Wolf Reik celia.x.aldacatalinas@gsk. com (C.A.-C.) [email protected]. uk (W.R.) Highlights Protocol for CRISPRa screens with single- cell readout to interrogate gene function Detailed description of CRISPRa screening procedures in mouse embryonic stem cells Detailed steps on how to construct derived single-cell sgRNA amplicon libraries CRISPR/Cas9 screens are a powerful approach to identify key regulators of biological processes. By combining pooled CRISPR/Cas9 screening with a single-cell RNA-sequencing readout, individual perturbations can be assessed in parallel both comprehensively and at scale. Importantly, this allows gene function and regulation to be interrogated at a cellular level in an unbiased manner. Here, we present a protocol to perform pooled CRISPR-activation screens in mouse embryonic stem cells using 103 Genomics scRNA-seq as a readout. Alda-Catalinas et al., STAR Protocols 2, 100426 June 18, 2021 ª 2021 The Authors. https://doi.org/10.1016/ j.xpro.2021.100426 ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas,1,4,7,* Melanie A. Eckersley-Maslin,1,5,6 and Wolf Reik1,2,3,8,* 1Epigenetics Programme, Babraham Institute, Cambridge CB22 3AT, UK 2Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK 3Centre for Trophoblast Research, University of -

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Spatiotemporal control of CRISPR/Cas9 gene editing Chenya Zhuo1, Jiabin Zhang1, Jung-Hwan Lee2, Ju Jiao3, Du Cheng4, Li Liu5, Hae-Won Kim2,YuTao1 and Mingqiang Li 1,6 The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/Cas9) gene editing technology, as a revolutionary breakthrough in genetic engineering, offers a promising platform to improve the treatment of various genetic and infectious diseases because of its simple design and powerful ability to edit different loci simultaneously. However, failure to conduct precise gene editing in specific tissues or cells within a certain time may result in undesirable consequences, such as serious off-target effects, representing a critical challenge for the clinical translation of the technology. Recently, some emerging strategies using genetic regulation, chemical and physical strategies to regulate the activity of CRISPR/Cas9 have shown promising results in the improvement of spatiotemporal controllability. Herein, in this review, we first summarize the latest progress of these advanced strategies involving cell-specific promoters, small-molecule activation and inhibition, bioresponsive delivery carriers, and optical/thermal/ultrasonic/magnetic activation. Next, we highlight the advantages and disadvantages of various strategies and discuss their obstacles and limitations in clinical translation. Finally, we propose viewpoints on directions that can be explored to -
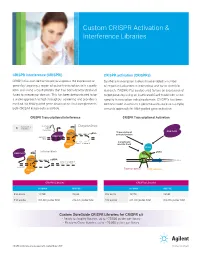
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

Synthetic CRISPR-Cas Gene Activators for Transcriptional Reprogramming in Bacteria
Corrected: Author correction ARTICLE DOI: 10.1038/s41467-018-04901-6 OPEN Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria Chen Dong1, Jason Fontana2, Anika Patel1, James M. Carothers 2,3,4 & Jesse G. Zalatan 1,2,4 Methods to regulate gene expression programs in bacterial cells are limited by the absence of effective gene activators. To address this challenge, we have developed synthetic bacterial transcriptional activators in E. coli by linking activation domains to programmable CRISPR-Cas 1234567890():,; DNA binding domains. Effective gene activation requires target sites situated in a narrow region just upstream of the transcription start site, in sharp contrast to the relatively flexible target site requirements for gene activation in eukaryotic cells. Together with existing tools for CRISPRi gene repression, these bacterial activators enable programmable control over multiple genes with simultaneous activation and repression. Further, the entire gene expression program can be switched on by inducing expression of the CRISPR-Cas system. This work will provide a foundation for engineering synthetic bacterial cellular devices with applications including diagnostics, therapeutics, and industrial biosynthesis. 1 Department of Chemistry, University of Washington, Seattle, WA 98195, USA. 2 Molecular Engineering & Sciences Institute, University of Washington, Seattle, WA 98195, USA. 3 Department of Chemical Engineering, University of Washington, Seattle, WA 98195, USA. 4 Center for Synthetic Biology, University of Washington, Seattle, WA 98195, USA. Correspondence and requests for materials should be addressed to J.M.C. (email: [email protected]) or to J.G.Z. (email: [email protected]) NATURE COMMUNICATIONS | (2018) 9:2489 | DOI: 10.1038/s41467-018-04901-6 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/s41467-018-04901-6 acteria are attractive targets for a wide variety of engi- in bacteria suggests that RpoZ may not be effective as a general Bneering applications. -

CRISPR-Cas9, Dickey-Wicker, and the Inner Space Race
Health Matrix: The Journal of Law- Medicine Volume 28 Issue 1 Article 9 2018 Shining City on a Hill at the Edge of Tomorrow: CRISPR-Cas9, Dickey-Wicker, and the Inner Space Race Zachary A. Zalewski Follow this and additional works at: https://scholarlycommons.law.case.edu/healthmatrix Part of the Health Law and Policy Commons Recommended Citation Zachary A. Zalewski, Shining City on a Hill at the Edge of Tomorrow: CRISPR-Cas9, Dickey-Wicker, and the Inner Space Race, 28 Health Matrix 469 (2018) Available at: https://scholarlycommons.law.case.edu/healthmatrix/vol28/iss1/9 This Note is brought to you for free and open access by the Student Journals at Case Western Reserve University School of Law Scholarly Commons. It has been accepted for inclusion in Health Matrix: The Journal of Law- Medicine by an authorized administrator of Case Western Reserve University School of Law Scholarly Commons. Health Matrix 28·Issue 1·2018 Shining City on a Hill at the Edge of Tomorrow: CRISPR-Cas9, Dickey-Wicker, and the Inner Space Race Zachary A. Zalewski† Contents Preface ................................................................................ 470 Introduction ........................................................................ 472 I. On Targeted Genome Editing: Towards Mastering (and Possibly Redesigning) the Blueprint of Life; a “Brief” History of CRISPR-Cas9 Development ......... 477 II. The CRISPR-Cas9 Gold Rush: Developments at Home and Abroad .................................................................. 485 A. The People’s Republic -

CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx Mori Embryonic Cells
insects Article CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells Wenbo Hu 1,2 , Xiaogang Wang 1, Sanyuan Ma 1,2, Zhangchuan Peng 1,2, Yang Cao 1,2 and Qingyou Xia 1,2,* 1 State Key Laboratory of Silkworm Genome Biology, Biological Science Research Center, Southwest University, Chongqing 400715, China; [email protected] (W.H.); [email protected] (X.W.); [email protected] (S.M.); [email protected] (Z.P.); [email protected] (Y.C.) 2 Chongqing Key Laboratory of Sericulture Science, Chongqing Engineering and Technology Research Center for Novel Silk Materials, Chongqing 400715, China * Correspondence: [email protected] Simple Summary: Based on a CRISPRa approach, activating endogenous fibroin heavy chain (FibH) gene expression in Bombyx mori embryonic (BmE) cells, which was driven by a combination of the dCas9-VPR (a tripartite activator, composed of VP64, p65, and Rta) and the sgRNA targeting to the promoter of FibH gene, was performed for investigating the biological roles of FibH in the development of silk gland cells. The activation of the endogenous FibH gene lead to up-regulation of cellular stress responses-related genes, which suggested a significant positive correlation between activated FibH gene expression and cellular stress responses. Moreover, the present findings might provide a potential model for studying the cellular stress responses caused by silk secretion disorder and lay a foundation for the understanding of silk gland development in silk-spinning insects. Citation: Hu, W.; Wang, X.; Ma, S.; Peng, Z.; Cao, Y.; Xia, Q. Abstract: The silkworm Bombyx mori is an economically important insect, as it is the main producer CRISPR-Mediated Endogenous of silk. -

The CRISPR-Cas System
Linköping University - Department of Physics, Chemistry and Biology Bachelor thesis, 16 hp - Educational program: Physics, Chemistry and Biology Spring term 2020 - LITH - IFM - G - EX -- 20/3902 -- SE The CRISPR-Cas system Can CRISPR bring about a brighter future? An extensive and up-to-date CRISPR overview, focusing on the CRISPR-Cas9 technology including a suggestion of a CRISPR-Cas9 laboratory assignment for University students. Sara Berggren, Isabella Enoksson and Cassandra Stens Examiner: Assoc. Prof. Lars-Göran Mårtensson 1 Abstract Derived from and inspired by the adaptive immune system of bacteria, CRISPR has gone from basic biology knowledge to a revolutionizing biotechnological tool, applicable in many research areas such as medicine, industry and agriculture. The full mechanism of CRISPR-Cas9 was first published in 2012 and various CRISPR-Cas systems have already passed the first stages of clinical trials as new gene therapies. The immense research has resulted in continuously growing knowledge of CRISPR systems and the technique seems to have the potential to greatly impact all life on our planet. Therefore, this literature study aims to thoroughly describe the CRISPR-Cas system, and further suggest an undergraduate laboratory exercise involving gene editing with the CRISPR-Cas9 tool. In this paper, we describe the fundamental technical background of the CRISPR-Cas system, especially emphasizing the most studied CRISPR-Cas9 system, its development and applications areas, as well as highlighting its current limitations and ethical concerns. The history of genetic engineering and the discovery of the CRISPR system is also described, along with a comparison with other established gene editing techniques. -

CRISPR/Cas9 Gene Editing
Oncology & Oncology & Immunology Immunology In Vivo Target Validation with KO and KI mice • from concept, disease models, to PK-PD-efficacy studies • Generation of cell lines with CRISPR-mediated gene knockout and knockin CRISPR/Cas9 Gene • Genome-wide screening for target identification and validation Timeline for CRISPR/CAS9 based method for knock In Service • Human and mouse CRISPR knockout libraries • Human CRISPR activation libraries 2-4 weeks 2-4 weeks 4-6 weeks 2-3 weeks 3-6 months Editing Platforms • Fast generation of GEMMs for genetic diseases sgRNA design & Microinjection Breeding, In vitro test & QC Genotyping preparation & Transplantation PK PD study F0 ssODN or vector construction (KI) CRISPR/Cas9 for target validation • Target validation in cells or mice in combination with other tools such as lentiviral gene overexpression, RNAi knockdown sgRNA for targeting • Phenotype ssODN for point mutation • Overexpression (Lenti) Engineered • Tumorigenesis • Knockdown (RNAi) cell lines • KO/KI (CRISPR) • Drug sensitivity Donor vector for replacement Target Target engagement In Vivo CRISPR Animal Models • Phenotype • Immune • from genome editing to disease models vs.Tumorigenesis KO/KI (CRISPR) Case study: Leptin and Leptin Receptor Knockout rats for obesity, Case study 2: DEN-induced Aldh2E487K KI mice could accelerate liver tumor GEMM • Tumor formation diabetes and fatty liver development • Drug sensitivity Case study: Gene knockout of gene X in MDA-MB-231 by CRISPR/Cas9 Lep-/- LepR -/- A gRNA #3 B WT WT Control 1 5 6 10 13 Gene X Glucose tolerance test #3-5 #3-1 Parental β-Actin Reference Gene X #3-8 #3-10 g3-6 1. Shengfang Jin et al., ALDH2(E487K) mutation increases protein turnover and (ECL) promotes murine hepatocarcinogenesis. -

Programmable and Portable CRISPR-Cas Transcriptional Activation in Bacteria
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.03.882431; this version posted January 3, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Programmable and portable CRISPR-Cas transcriptional activation in bacteria Hsing-I Ho1, Jennifer Fang2, Jacky Cheung3, Harris H. Wang1,4* Affiliations 1Department of Systems Biology, Columbia University, New York, NY, USA. 2Department of Biology, Columbia University, New York, NY, USA. 3Department of Computer Science and Biology, Columbia University, New York, NY, USA 4Department of Pathology and Cell Biology, Columbia University, New York, NY, USA *Correspondence to: [email protected] ABSTRACT Programmable gene activation enables fine-tuned regulation of endogenous and synthetic gene circuits to control cellular behavior. While CRISPR-Cas-mediated gene activation have been extensively developed for eukaryotic systems, similar strategies have been difficult to implement in bacteria. Here, we present a generalizable platform for screening and selection of functional bacterial CRISPR-Cas transcription activators. Using this platform, we identified a novel CRISPR activator, dCas9-AsiA, that could activate gene expression by up to 200-fold across genomic and plasmid targets with diverse promoters after directed evolution. The evolved dCas9-AsiA can simultaneously mediate activation and repression of bacterial regulons in E. coli. We further identified hundreds of promoters with varying basal expression that could be induced by dCas9-AsiA, which provides a rich resource of genetic parts for inducible gene activation. -

CRISPR/Dcas9-Based Systems: Mechanisms and Applications in Plant Sciences
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 1 February 2021 doi:10.20944/preprints202102.0033.v1 CRISPR/dCas9-based systems: Mechanisms and applications in plant sciences Chou Khai Soong Karlsona, Siti Nurfadhlina Mohd Noora, Nadja Nolteb, and Boon Chin Tana,* a Center for Research in Biotechnology for Agriculture (CEBAR), University of Malaya, 50603 Kuala Lumpur, Malaysia b Department of Plant Sciences, Wageningen University & Research, Droevendaalsesteeg 4, 6708 PB Wageningen, The Netherlands *Corresponding Author: Email address: [email protected]; Tel: +603-7967 7982 Abstract RNA-guided genomic transcriptional regulation tools, namely Clustered Regularly Interspaced Short Palindromic Repeats interference (CRISPRi) and CRISPR-mediated gene activation (CRISPRa), are a powerful technology for the field of functional genomics. Deriving from the CRISPR/Cas9 system, both systems comprise a catalytically dead Cas9 (dCas9) and a single guide RNA (sgRNA). This type of dCas9 is incapable of cleaving DNA but retains its ability to specifically bind to DNA. The binding of the dCas9/sgRNA complex to a target gene results in transcriptional interference. The CRISPR/dCas9 system has been explored as a tool for transcriptional modulation and genome imaging. Despite its potential applications and benefits, the challenges and limitations faced by the CRISPR/dCas9 system include the off-target effects, PAM sequence requirement, efficient delivery methods, and the CRISPR/dCas9-interfered crops being labeled as genetically modified organisms in several countries. This review highlights the progression of CRISPR/dCas9 technology as well as its applications and potential challenges in crop improvement. Keywords: CRISPR interference; CRISPR/dCas9 system; crop improvement; gene silencing; RNAi; transcriptional regulation Introduction Recent advancements in omics-based technologies and bioinformatics methodologies have facilitated the ever-growing field of functional genomics and system biology (Guigo & De Hoon, 2018; Yadav & Tripathi, 2018).