Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/Dcas9 Activator Complex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas, Melanie A. Eckersley-Maslin, Wolf Reik celia.x.aldacatalinas@gsk. com (C.A.-C.) [email protected]. uk (W.R.) Highlights Protocol for CRISPRa screens with single- cell readout to interrogate gene function Detailed description of CRISPRa screening procedures in mouse embryonic stem cells Detailed steps on how to construct derived single-cell sgRNA amplicon libraries CRISPR/Cas9 screens are a powerful approach to identify key regulators of biological processes. By combining pooled CRISPR/Cas9 screening with a single-cell RNA-sequencing readout, individual perturbations can be assessed in parallel both comprehensively and at scale. Importantly, this allows gene function and regulation to be interrogated at a cellular level in an unbiased manner. Here, we present a protocol to perform pooled CRISPR-activation screens in mouse embryonic stem cells using 103 Genomics scRNA-seq as a readout. Alda-Catalinas et al., STAR Protocols 2, 100426 June 18, 2021 ª 2021 The Authors. https://doi.org/10.1016/ j.xpro.2021.100426 ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas,1,4,7,* Melanie A. Eckersley-Maslin,1,5,6 and Wolf Reik1,2,3,8,* 1Epigenetics Programme, Babraham Institute, Cambridge CB22 3AT, UK 2Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK 3Centre for Trophoblast Research, University of -

Interfering with Retrotransposition by Two Types of CRISPR Effectors
Zhang et al. Cell Discovery (2020) 6:30 Cell Discovery https://doi.org/10.1038/s41421-020-0164-0 www.nature.com/celldisc ARTICLE Open Access Interfering with retrotransposition by two types of CRISPR effectors: Cas12a and Cas13a Niubing Zhang1,2, Xinyun Jing1, Yuanhua Liu3, Minjie Chen1,2, Xianfeng Zhu2, Jing Jiang2, Hongbing Wang4, Xuan Li1 and Pei Hao3 Abstract CRISPRs are a promising tool being explored in combating exogenous retroviral pathogens and in disabling endogenous retroviruses for organ transplantation. The Cas12a and Cas13a systems offer novel mechanisms of CRISPR actions that have not been evaluated for retrovirus interference. Particularly, a latest study revealed that the activated Cas13a provided bacterial hosts with a “passive protection” mechanism to defend against DNA phage infection by inducing cell growth arrest in infected cells, which is especially significant as it endows Cas13a, a RNA-targeting CRISPR effector, with mount defense against both RNA and DNA invaders. Here, by refitting long terminal repeat retrotransposon Tf1 as a model system, which shares common features with retrovirus regarding their replication mechanism and life cycle, we repurposed CRISPR-Cas12a and -Cas13a to interfere with Tf1 retrotransposition, and evaluated their different mechanisms of action. Cas12a exhibited strong inhibition on retrotransposition, allowing marginal Tf1 transposition that was likely the result of a lasting pool of Tf1 RNA/cDNA intermediates protected within virus-like particles. The residual activities, however, were completely eliminated with new constructs for persistent crRNA targeting. On the other hand, targeting Cas13a to Tf1 RNA intermediates significantly inhibited Tf1 retrotransposition. However, unlike in bacterial hosts, the sustained activation of Cas13a by Tf1 transcripts did not cause cell growth arrest in S. -

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Complete Article
The EMBO Journal Vol. I No. 12 pp. 1539-1544, 1982 Long terminal repeat-like elements flank a human immunoglobulin epsilon pseudogene that lacks introns Shintaro Ueda', Sumiko Nakai, Yasuyoshi Nishida, lack the entire IVS have been found in the gene families of the Hiroshi Hisajima, and Tasuku Honjo* mouse a-globin (Nishioka et al., 1980; Vanin et al., 1980), the lambda chain (Hollis et al., 1982), Department of Genetics, Osaka University Medical School, Osaka 530, human immunoglobulin Japan and the human ,B-tubulin (Wilde et al., 1982a, 1982b). The mouse a-globin processed gene is flanked by long terminal Communicated by K.Rajewsky Received on 30 September 1982 repeats (LTRs) of retrovirus-like intracisternal A particles on both sides, although their orientation is opposite to each There are at least three immunoglobulin epsilon genes (C,1, other (Lueders et al., 1982). The human processed genes CE2, and CE) in the human genome. The nucleotide sequences described above have poly(A)-like tails -20 bases 3' to the of the expressed epsilon gene (CE,) and one (CE) of the two putative poly(A) addition signal and are flanked by direct epsilon pseudogenes were compared. The results show that repeats of several bases on both sides (Hollis et al., 1982; the CE3 gene lacks the three intervening sequences entirely and Wilde et al., 1982a, 1982b). Such direct repeats, which were has a 31-base A-rich sequence 16 bases 3' to the putative also found in human small nuclear RNA pseudogenes poly(A) addition signal, indicating that the CE3 gene is a pro- (Arsdell et al., 1981), might have been formed by repair of cessed gene. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

Retroviral Insertions Into a Herpesvirus Are Clustered At
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 3855-3859, May 1993 Biochemistry Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences (Marek disease virus/reticuloendotheliosis virus/long terminal repeat/insertional mutagenesis/retroviral integration) DAN JONES*, ROBERT ISFORTt, RICHARD WITTERt, RHONDA KoST*, AND HSING-JIEN KUNG*§ *Department of Molecular Biology and Microbiology, Case Western Reserve University, Cleveland, OH 44106; tGenetic Toxicology Section, Human and Environmental Safety Division, The Procter and Gamble Company, Miami Valley Laboratories, Cincinnati, OH 45239; and tU.S. Department of Agriculture Agricultural Research Service, Avian Disease and Oncology Laboratory, 3606 East Mount Hope, East Lansing, MI 48823 Communicated by Frederick C. Robbins, January 11, 1993 (receivedfor review November 3, 1992) ABSTRACT We previously described the integration of a integrations are associated with large structural changes in nonacute retrovirus, reticuloendotheliosis virus (REV), into the MDV genome in both the Us and Rs regions (11). the genome of a herpesvirus, Marek disease virus (MDV), By coinfection of duck embryo fibroblasts (DEFs) with following both long-term and short-term coinfection in cul- both REV and MDV, we demonstrated that this process of tured fibroblasts. The long-term coinfection occurred in the retroviral integration can also occur within several passages course of attenuating the oncogenicity ofthe JM strain ofMDV of cocultivation (10). The structure of the inserted REV and was sustained for >100 passages. The short-term coinfec- sequences resembles those of cellular integrated proviruses, tion, which spanned only 16 passages, was designed to recreate with loss of the terminal nucleotides of the retrovirus and a the insertion phenomenon under controlled conditions. -

Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Spatiotemporal control of CRISPR/Cas9 gene editing Chenya Zhuo1, Jiabin Zhang1, Jung-Hwan Lee2, Ju Jiao3, Du Cheng4, Li Liu5, Hae-Won Kim2,YuTao1 and Mingqiang Li 1,6 The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/Cas9) gene editing technology, as a revolutionary breakthrough in genetic engineering, offers a promising platform to improve the treatment of various genetic and infectious diseases because of its simple design and powerful ability to edit different loci simultaneously. However, failure to conduct precise gene editing in specific tissues or cells within a certain time may result in undesirable consequences, such as serious off-target effects, representing a critical challenge for the clinical translation of the technology. Recently, some emerging strategies using genetic regulation, chemical and physical strategies to regulate the activity of CRISPR/Cas9 have shown promising results in the improvement of spatiotemporal controllability. Herein, in this review, we first summarize the latest progress of these advanced strategies involving cell-specific promoters, small-molecule activation and inhibition, bioresponsive delivery carriers, and optical/thermal/ultrasonic/magnetic activation. Next, we highlight the advantages and disadvantages of various strategies and discuss their obstacles and limitations in clinical translation. Finally, we propose viewpoints on directions that can be explored to -

Human Immunodeficiency Virus Type 1 Two-Long Terminal Repeat
Isabel Olivares, et al.: HIV-1 Two-Long Terminal Repeat Circles Contents available at PubMed www.aidsreviews.com PERMANYER AIDS Rev. 2016;18:23-31 www.permanyer.com Human Immunodeficiency Virus Type 1 Two-Long Terminal Repeat Circles: A Subject for Debate Isabel Olivares, María Pernas, Concepción Casado and Cecilio López-Galindez Department of Molecular Virology, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Majadahonda, Madrid, Spain © Permanyer Publications 2016 Abstract . HIV-1 infections are characterized by the integration of the reverse transcribed genomic RNA into the host chromosomes making up the provirus. In addition to the integrated proviral DNA, there are other forms of linear and circular unintegrated viral DNA in HIV-1-infected cells. One of these forms, known as two-long terminal repeat circles, has been extensively studied and characterized both in in vitro infected cells and in cells of the publisher from patients. Detection of two-long terminal repeat circles has been proposed as a marker of antire troviral treatment efficacy or ongoing replication in patients with undetectable viral load. But not all authors agree with this use because of the uncertainty about the lifespan of the two-long terminal repeat circles. We review the major studies estimating the half-life of the two-long terminal repeat circles as well as those proposing its detection as a marker of ongoing replication or therapeutic efficacy. We also review the charac- teristic of these circular forms and the difficulties in its detection and quantification. The variety of approaches and methods used in the two-long terminal repeat quantification as well as the low reliability of some methods make the comparison between results difficult. -

Repetitive Elements in Humans
International Journal of Molecular Sciences Review Repetitive Elements in Humans Thomas Liehr Institute of Human Genetics, Jena University Hospital, Friedrich Schiller University, Am Klinikum 1, D-07747 Jena, Germany; [email protected] Abstract: Repetitive DNA in humans is still widely considered to be meaningless, and variations within this part of the genome are generally considered to be harmless to the carrier. In contrast, for euchromatic variation, one becomes more careful in classifying inter-individual differences as meaningless and rather tends to see them as possible influencers of the so-called ‘genetic background’, being able to at least potentially influence disease susceptibilities. Here, the known ‘bad boys’ among repetitive DNAs are reviewed. Variable numbers of tandem repeats (VNTRs = micro- and minisatellites), small-scale repetitive elements (SSREs) and even chromosomal heteromorphisms (CHs) may therefore have direct or indirect influences on human diseases and susceptibilities. Summarizing this specific aspect here for the first time should contribute to stimulating more research on human repetitive DNA. It should also become clear that these kinds of studies must be done at all available levels of resolution, i.e., from the base pair to chromosomal level and, importantly, the epigenetic level, as well. Keywords: variable numbers of tandem repeats (VNTRs); microsatellites; minisatellites; small-scale repetitive elements (SSREs); chromosomal heteromorphisms (CHs); higher-order repeat (HOR); retroviral DNA 1. Introduction Citation: Liehr, T. Repetitive In humans, like in other higher species, the genome of one individual never looks 100% Elements in Humans. Int. J. Mol. Sci. alike to another one [1], even among those of the same gender or between monozygotic 2021, 22, 2072. -
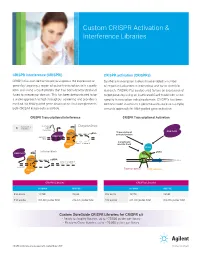
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

Characterization of the Long Terminal Repeat of the Endogenous
www.nature.com/scientificreports OPEN Characterization of the Long Terminal Repeat of the Endogenous Retrovirus-derived microRNAs in Received: 31 May 2018 Accepted: 12 September 2019 the Olive Flounder Published: xx xx xxxx Hee-Eun Lee1,2, Ara Jo1,2, Jennifer Im1,2, Hee-Jae Cha 3, Woo-Jin Kim4, Hyun Hee Kim5,6, Dong-Soo Kim7, Won Kim8, Tae-Jin Yang 9 & Heui-Soo Kim2,10 Endogenous retroviruses (ERVs) have been identifed at diferent copy numbers in various organisms. The long terminal repeat (LTR) element of an ERV has the capacity to exert regulatory infuence as both a promoter and enhancer of cellular genes. Here, we describe olive founder (OF)-ERV9, derived from chromosome 9 of the olive founder. OF-ERV9-LTR provide binding sites for various transcription factors and showed enhancer activity. The OF-ERV9-LTR demonstrates high sequence similarity with the 3′ untranslated region (UTR) of various genes that also contain seed sequences (TGTTTTG) that bind the LTR-derived microRNA(miRNA), OF-miRNA-307. Additionally, OF-miRNA-307 collaborates with transcription factors located in OF-ERV9-LTR to regulate gene expression. Taken together, our data facilitates a greater understanding of the molecular function of OF-ERV families and suggests that OF- miRNA-307 may act as a super-enhancer miRNA regulating gene activity. Paralichthys olivaceus, known as olive founder (OF) is an economically important marine fatfsh which is exten- sively cultured in Korea, China and Japan. Due to their high economic value, there are several selective breed- ing programs in place, such as those involving sex manipulation, owing to diferences in growth speed and size between male and female olive founders1–3. -

A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
METHODS, MODELS & TECHNIQUES Prospects & Overviews www.bioessays-journal.com A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities Yang Liu and Baojun Wang* friend or foe” system, scientists are CRISPR (clustered regularly interspaced short palindromic repeats) activation able to guide the endonucleases to (CRISPRa) in bacteria is an attractive method for programmable gene their desired DNA or RNA targets.[5–8] activation. Recently, a eukaryote-like, 54-dependent CRISPRa system has CRISPR regulation relies on inactivated been reported. It exhibits high dynamic ranges and permits flexible target site CRISPR endonucleases. The nuclease- deficient CRISPR DNA endonucleases, selection. Here, an overview of the existing strategies of CRISPRa in bacteria for instances dCas9 and ddCpf1 (dCas12), is presented, and the characteristics and design principles of the CRISPRa are effectively programmable DNA bind- system are introduced. Possible scenarios for applying the eukaryote-like ing domains. When these domains are CRISPRa system is discussed with corresponding suggestions for tethered to transactivation domains or performance optimization and future functional expansion. The authors subunits of RNA polymerase, they ac- envision the new eukaryote-like CRISPRa system enabling novel designs in tivate the promoters near the CRISPR target sites. This strategy has been 54 multiplexed gene regulation and promoting research in the -dependent widely utilized in both eukaryotes and gene regulatory networks among a variety of biotechnology relevant or prokaryotes,[9–17] particularly in the former, disease-associated bacterial species. where the transcription activation mecha- nisms and the activators are well-studied. While CRISPRa in eukaryotes enjoys much success and is continuously im- 1.