CRISPR/Dcas9-Based Systems: Mechanisms and Applications in Plant Sciences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pooled CRISPR-Activation Screening Coupled with Single-Cell RNA-Seq in Mouse Embryonic Stem Cells
ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas, Melanie A. Eckersley-Maslin, Wolf Reik celia.x.aldacatalinas@gsk. com (C.A.-C.) [email protected]. uk (W.R.) Highlights Protocol for CRISPRa screens with single- cell readout to interrogate gene function Detailed description of CRISPRa screening procedures in mouse embryonic stem cells Detailed steps on how to construct derived single-cell sgRNA amplicon libraries CRISPR/Cas9 screens are a powerful approach to identify key regulators of biological processes. By combining pooled CRISPR/Cas9 screening with a single-cell RNA-sequencing readout, individual perturbations can be assessed in parallel both comprehensively and at scale. Importantly, this allows gene function and regulation to be interrogated at a cellular level in an unbiased manner. Here, we present a protocol to perform pooled CRISPR-activation screens in mouse embryonic stem cells using 103 Genomics scRNA-seq as a readout. Alda-Catalinas et al., STAR Protocols 2, 100426 June 18, 2021 ª 2021 The Authors. https://doi.org/10.1016/ j.xpro.2021.100426 ll OPEN ACCESS Protocol Pooled CRISPR-activation screening coupled with single-cell RNA-seq in mouse embryonic stem cells Celia Alda-Catalinas,1,4,7,* Melanie A. Eckersley-Maslin,1,5,6 and Wolf Reik1,2,3,8,* 1Epigenetics Programme, Babraham Institute, Cambridge CB22 3AT, UK 2Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK 3Centre for Trophoblast Research, University of -

A CRISPR Activation and Interference Toolkit for Industrial Saccharomyces Cerevisiae Strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård*
www.nature.com/scientificreports OPEN A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6‑12 Elena Cámara, Ibai Lenitz & Yvonne Nygård* Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology‑directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fuorescent protein‑encoding genes under two diferent promoters allowing expression alterations up to ~ 2.5‑fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design‑build‑test cycle for developing various industrial production strains. Te yeast Saccharomyces cerevisiae is one of the most commonly used microorganisms for industrial applications ranging from wine and beer fermentations to the production of biofuels and high-value metabolites1,2. How- ever, some of the current production processes are compromised by low yields and productivities, thus further optimization is required3. -

Advances in Genomics for Drug Development
G C A T T A C G G C A T genes Review Advances in Genomics for Drug Development Roberto Spreafico , Leah B. Soriaga, Johannes Grosse, Herbert W. Virgin and Amalio Telenti * Vir Biotechnology, Inc., San Francisco, CA 94158, USA; Rspreafi[email protected] (R.S.); [email protected] (L.B.S.); [email protected] (J.G.); [email protected] (H.W.V.) * Correspondence: [email protected] Received: 24 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Drug development (target identification, advancing drug leads to candidates for preclinical and clinical studies) can be facilitated by genetic and genomic knowledge. Here, we review the contribution of population genomics to target identification, the value of bulk and single cell gene expression analysis for understanding the biological relevance of a drug target, and genome-wide CRISPR editing for the prioritization of drug targets. In genomics, we discuss the different scope of genome-wide association studies using genotyping arrays, versus exome and whole genome sequencing. In transcriptomics, we discuss the information from drug perturbation and the selection of biomarkers. For CRISPR screens, we discuss target discovery, mechanism of action and the concept of gene to drug mapping. Harnessing genetic support increases the probability of drug developability and approval. Keywords: druggability; loss-of-function; CRISPR 1. Introduction For over 20 years, genomics has been used as a tool for accelerating drug development. Various conceptual approaches and techniques assist target identification, target prioritization and tractability, as well as the prediction of outcomes from pharmacological perturbations. These basic premises are now supported by a rapid expansion of population genomics initiatives (sequencing or genotyping of hundreds of thousands of individuals), in-depth understanding of disease and drug perturbation at the tissue and single-cell level as measured by transcriptome analysis, and by the capacity to screen for loss of function or activation of genes, genome-wide, using CRISPR technologies. -

Spatiotemporal Control of CRISPR/Cas9 Gene Editing
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN Spatiotemporal control of CRISPR/Cas9 gene editing Chenya Zhuo1, Jiabin Zhang1, Jung-Hwan Lee2, Ju Jiao3, Du Cheng4, Li Liu5, Hae-Won Kim2,YuTao1 and Mingqiang Li 1,6 The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/Cas9) gene editing technology, as a revolutionary breakthrough in genetic engineering, offers a promising platform to improve the treatment of various genetic and infectious diseases because of its simple design and powerful ability to edit different loci simultaneously. However, failure to conduct precise gene editing in specific tissues or cells within a certain time may result in undesirable consequences, such as serious off-target effects, representing a critical challenge for the clinical translation of the technology. Recently, some emerging strategies using genetic regulation, chemical and physical strategies to regulate the activity of CRISPR/Cas9 have shown promising results in the improvement of spatiotemporal controllability. Herein, in this review, we first summarize the latest progress of these advanced strategies involving cell-specific promoters, small-molecule activation and inhibition, bioresponsive delivery carriers, and optical/thermal/ultrasonic/magnetic activation. Next, we highlight the advantages and disadvantages of various strategies and discuss their obstacles and limitations in clinical translation. Finally, we propose viewpoints on directions that can be explored to -

Quantitative CRISPR Interference Screens in Yeast Identify Chemical-Genetic Interactions and New Rules for Guide RNA Design Justin D
Smith et al. Genome Biology (2016) 17:45 DOI 10.1186/s13059-016-0900-9 RESEARCH Open Access Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design Justin D. Smith1,2, Sundari Suresh1, Ulrich Schlecht1, Manhong Wu3, Omar Wagih4, Gary Peltz3, Ronald W. Davis1, Lars M. Steinmetz1,2,5, Leopold Parts2,5,6* and Robert P. St.Onge1* Abstract Background: Genome-scale CRISPR interference (CRISPRi) has been used in human cell lines; however, the features of effective guide RNAs (gRNAs) in different organisms have not been well characterized. Here, we define rules that determine gRNA effectiveness for transcriptional repression in Saccharomyces cerevisiae. Results: We create an inducible single plasmid CRISPRi system for gene repression in yeast, and use it to analyze fitness effects of gRNAs under 18 small molecule treatments. Our approach correctly identifies previously described chemical-genetic interactions, as well as a new mechanism of suppressing fluconazole toxicity by repression of the ERG25 gene. Assessment of multiple target loci across treatments using gRNA libraries allows us to determine generalizable features associated with gRNA efficacy. Guides that target regions with low nucleosome occupancy and high chromatin accessibility are clearly more effective. We also find that the best region to target gRNAs is between the transcription start site (TSS) and 200 bp upstream of the TSS. Finally, unlike nuclease-proficient Cas9 in human cells, the specificity of truncated gRNAs (18 nt of complementarity to the target) is not clearly superior to full-length gRNAs (20 nt of complementarity), as truncated gRNAs are generally less potent against both mismatched and perfectly matched targets. -

CRISPR-Cas Systems Restrict Horizontal Gene Transfer In
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.19.304717; this version posted September 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CRISPR-Cas systems restrict horizontal gene transfer in 2 Pseudomonas aeruginosa 3 Running title: CRISPR-Cas systems in Pseudomonas aeruginosa 4 5 Authors: Rachel M. Wheatley1,* and R. Craig MacLean1 6 7 Affiliations: Department of Zoology, University of Oxford, Oxford OX1 3PS, UK 8 9 Contact details: [email protected] 10 11 Competing interests 12 This project was supported by Wellcome Trust Grant 106918/Z/15/Z held by RCM. The authors declare 13 they have no conflict of interest. 14 15 16 17 18 19 20 21 22 23 24 25 26 27 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.19.304717; this version posted September 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 28 Abstract 29 CRISPR-Cas systems provide bacteria and archaea with an adaptive immune system that targets foreign 30 DNA. However, the xenogenic nature of immunity provided by CRISPR-Cas raises the possibility that 31 these systems may constrain horizontal gene transfer. Here we test this hypothesis in the opportunistic 32 pathogen Pseudomonas aeruginosa, which has emerged an important model system for understanding 33 CRISPR-Cas function. Across the diversity of P. aeruginosa, active CRISPR-Cas systems are 34 associated with smaller genomes and a reduced GC content, suggesting that CRISPR-Cas inhibits the 35 acquisition of foreign DNA. -
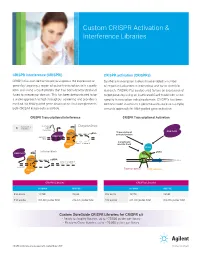
Custom CRISPR Activation & Interference Libraries
Custom CRISPR Activation & Interference Libraries CRISPR interference (CRISPRi) CRISPR activation (CRISPRa) CRISPR/Cas can be harnessed to suppress the expression of Synthetic transcription factors have enabled a number genes by targeting a region of active transcription with a guide of important advances in biomedical and basic scientific RNA and using a Cas9 protein that has been deactivated and research. CRISPR/Cas can be used to turn on expression of fused to a repressor domain. This has been demonstrated to be target genes by using an inactivated Cas9 fused with a non- a viable approach to high throughput screening and provides a specific transcription inducing domain. CRISPRa has been method for RNA-guided gene deactivation that complements demonstrated to work on a genome-wide scale as a simple, both CRISPR knock-outs and RNAi. versatile approach for RNA-guided gene activation. CRISPR Transcriptional Interference CRISPR Transcriptional Activation inactivating interrupted Elongation Block mutations mRNA x transcript RNA Pol II sgRNA Transcriptional activator protein genomic RNA Pol II DNA Catalytically dCAS9 Exon 1 of gene X inactive Cas9 VP64 Initiation Block gRNA RNA Pol II dCAS9 x sgRNA Gene of interest Exon 1 of gene X promoter Target sequence dCAS9 PAM sequence CRISPRi Libraries CRISPRa Libraries HUMAN MOUSE HUMAN MOUSE # of Genes 18,730 19,846 # of Genes 18,574 19,949 # of Guides 205,648 guides total 212,376 guides total # of Guides 201,530 guides total 208,066 guides total Custom SureGuide CRISPR Libraries for CRISPR a/i • Ready-to-Amplify libraries, up to ~70,000 guides per library • Ready-to-Clone libraries, up to ~70,000 guides per library CRISPR a/i libraries are designed with content from UCSF. -

A Novel Eukaryote‐Like CRISPR Activation Tool in Bacteria
METHODS, MODELS & TECHNIQUES Prospects & Overviews www.bioessays-journal.com A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities Yang Liu and Baojun Wang* friend or foe” system, scientists are CRISPR (clustered regularly interspaced short palindromic repeats) activation able to guide the endonucleases to (CRISPRa) in bacteria is an attractive method for programmable gene their desired DNA or RNA targets.[5–8] activation. Recently, a eukaryote-like, 54-dependent CRISPRa system has CRISPR regulation relies on inactivated been reported. It exhibits high dynamic ranges and permits flexible target site CRISPR endonucleases. The nuclease- deficient CRISPR DNA endonucleases, selection. Here, an overview of the existing strategies of CRISPRa in bacteria for instances dCas9 and ddCpf1 (dCas12), is presented, and the characteristics and design principles of the CRISPRa are effectively programmable DNA bind- system are introduced. Possible scenarios for applying the eukaryote-like ing domains. When these domains are CRISPRa system is discussed with corresponding suggestions for tethered to transactivation domains or performance optimization and future functional expansion. The authors subunits of RNA polymerase, they ac- envision the new eukaryote-like CRISPRa system enabling novel designs in tivate the promoters near the CRISPR target sites. This strategy has been 54 multiplexed gene regulation and promoting research in the -dependent widely utilized in both eukaryotes and gene regulatory networks among a variety of biotechnology relevant or prokaryotes,[9–17] particularly in the former, disease-associated bacterial species. where the transcription activation mecha- nisms and the activators are well-studied. While CRISPRa in eukaryotes enjoys much success and is continuously im- 1. -

CRISPR Interference (Crispri) for Gene Regulation and Succinate Production in Cyanobacterium S
Huang et al. Microb Cell Fact (2016) 15:196 DOI 10.1186/s12934-016-0595-3 Microbial Cell Factories RESEARCH Open Access CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942 Chun‑Hung Huang, Claire R. Shen, Hung Li, Li‑Yu Sung, Meng‑Ying Wu and Yu‑Chen Hu* Abstract Background: Cyanobacterium Synechococcus elongatus PCC 7942 holds promise for biochemical conversion, but gene deletion in PCC 7942 is time-consuming and may be lethal to cells. CRISPR interference (CRISPRi) is an emerging technology that exploits the catalytically inactive Cas9 (dCas9) and single guide RNA (sgRNA) to repress sequence- specific genes without the need of gene knockout, and is repurposed to rewire metabolic networks in various pro‑ caryotic cells. Results: To employ CRISPRi for the manipulation of gene network in PCC 7942, we integrated the cassettes express‑ ing enhanced yellow fluorescent protein (EYFP), dCas9 and sgRNA targeting different regions on eyfp into the PCC 7942 chromosome. Co-expression of dCas9 and sgRNA conferred effective and stable suppression of EYFP produc‑ tion at efficiencies exceeding 99%, without impairing cell growth. We next integrated the dCas9 and sgRNA targeting endogenous genes essential for glycogen accumulation (glgc) and succinate conversion to fumarate (sdhA and sdhB). Transcription levels of glgc, sdhA and sdhB were effectively suppressed with efficiencies depending on the sgRNA binding site. Targeted suppression of glgc reduced the expression to 6.2%, attenuated the glycogen accumulation to 4.8% and significantly enhanced the succinate titer. Targeting sdhA or sdhB also effectively downregulated the gene expression and enhanced the succinate titer 12.5-fold to 0.58–0.63 mg/L. -

RNA Interference in the Age of CRISPR: Will CRISPR Interfere with Rnai?
International Journal of Molecular Sciences Review RNA Interference in the Age of CRISPR: Will CRISPR Interfere with RNAi? Unnikrishnan Unniyampurath 1, Rajendra Pilankatta 2 and Manoj N. Krishnan 1,* 1 Program on Emerging Infectious Diseases, Duke-NUS Medical School, 8 College Road, Singapore 169857, Singapore; [email protected] 2 Department of Biochemistry and Molecular Biology, School of Biological Sciences, Central University of Kerala, Nileshwar 671328, India; [email protected] * Correspondence: [email protected]; Tel.: +65-6516-2666 Academic Editor: Michael Ladomery Received: 2 November 2015; Accepted: 15 February 2016; Published: 26 February 2016 Abstract: The recent emergence of multiple technologies for modifying gene structure has revolutionized mammalian biomedical research and enhanced the promises of gene therapy. Over the past decade, RNA interference (RNAi) based technologies widely dominated various research applications involving experimental modulation of gene expression at the post-transcriptional level. Recently, a new gene editing technology, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and the CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) system, has received unprecedented acceptance in the scientific community for a variety of genetic applications. Unlike RNAi, the CRISPR/Cas9 system is bestowed with the ability to introduce heritable precision insertions and deletions in the eukaryotic genome. The combination of popularity and superior capabilities of CRISPR/Cas9 system raises the possibility that this technology may occupy the roles currently served by RNAi and may even make RNAi obsolete. We performed a comparative analysis of the technical aspects and applications of the CRISPR/Cas9 system and RNAi in mammalian systems, with the purpose of charting out a predictive picture on whether the CRISPR/Cas9 system will eclipse the existence and future of RNAi. -

Development of CRISPR Interference (Crispri) Platform for Metabolic Engineering of Leuconostoc Citreum and Its Application for Engineering Riboflavin Biosynthesis
International Journal of Molecular Sciences Article Development of CRISPR Interference (CRISPRi) Platform for Metabolic Engineering of Leuconostoc citreum and Its Application for Engineering Riboflavin Biosynthesis Jaewoo Son 1, Seung Hoon Jang 1, Ji Won Cha 1 and Ki Jun Jeong 1,2,* 1 Department of Chemical and Biomolecular Engineering, BK21 Plus program, KAIST, 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea; [email protected] (J.S.); [email protected] (S.H.J.); [email protected] (J.W.C.) 2 Institute for The BioCentury, KAIST, 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Korea * Correspondence: [email protected]; Tel.: +82-42-350-3934 Received: 1 July 2020; Accepted: 3 August 2020; Published: 5 August 2020 Abstract: Leuconostoc citreum, a hetero-fermentative type of lactic acid bacteria, is a crucial probiotic candidate because of its ability to promote human health. However, inefficient gene manipulation tools limit its utilization in bioindustries. We report, for the first time, the development of a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) interference (CRISPRi) system for engineering L. citreum. For reliable expression, the expression system of synthetic single guide RNA (sgRNA) and the deactivated Cas9 of Streptococcus pyogenes (SpdCas9) were constructed in a bicistronic design (BCD) platform using a high-copy-number plasmid. The expression of SpdCas9 and sgRNA was optimized by examining the combination of two synthetic promoters and Shine–Dalgarno sequences; the strong expression of sgRNA and the weak expression of SpdCas9 exhibited the most significant downregulation (20-fold decrease) of the target gene (sfGFP), without cell growth retardation caused by SpdCas9 overexpression. -

Challenges, New Technologies and Their Use in Plants
International Journal of Molecular Sciences Review Chromatin Manipulation and Editing: Challenges, New Technologies and Their Use in Plants Kateryna Fal 1,† , Denisa Tomkova 2,†, Gilles Vachon 1 , Marie-Edith Chabouté 2, Alexandre Berr 2,* and Cristel C. Carles 1,* 1 Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS, CEA, INRAE, IRIG-LPCV, 38000 Grenoble, France; [email protected] (K.F.); [email protected] (G.V.) 2 Institut de Biologie Moléculaire des Plantes du CNRS, Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg CEDEX, France; [email protected] (D.T.); [email protected] (M.-E.C.) * Correspondence: [email protected] (A.B.); [email protected] (C.C.C.) † These authors contributed equally to this work. Abstract: An ongoing challenge in functional epigenomics is to develop tools for precise manipulation of epigenetic marks. These tools would allow moving from correlation-based to causal-based findings, a necessary step to reach conclusions on mechanistic principles. In this review, we describe and discuss the advantages and limits of tools and technologies developed to impact epigenetic marks, and which could be employed to study their direct effect on nuclear and chromatin structure, on transcription, and their further genuine role in plant cell fate and development. On one hand, epigenome-wide approaches include drug inhibitors for chromatin modifiers or readers, nanobodies against histone marks or lines expressing modified histones or mutant chromatin effectors. On the other hand, locus-specific approaches consist in targeting precise regions on the chromatin, with engineered proteins able to modify epigenetic marks.