BIO-Annual-Report-2003.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Analysis of the Homeobox Gene Tur-2 During Mouse Embryogenesis
Functional Analysis of The Homeobox Gene Tur-2 During Mouse Embryogenesis Shao Jun Tang A thesis submitted in conformity with the requirements for the Degree of Doctor of Philosophy Graduate Department of Molecular and Medical Genetics University of Toronto March, 1998 Copyright by Shao Jun Tang (1998) National Library Bibriothèque nationale du Canada Acquisitions and Acquisitions et Bibiiographic Services seMces bibliographiques 395 Wellington Street 395, rue Weifington OtbawaON K1AW OttawaON KYAON4 Canada Canada The author has granted a non- L'auteur a accordé une licence non exclusive licence alIowing the exclusive permettant à la National Library of Canada to Bibliothèque nationale du Canada de reproduce, loan, distri%uteor sell reproduire, prêter' distribuer ou copies of this thesis in microform, vendre des copies de cette thèse sous paper or electronic formats. la forme de microfiche/nlm, de reproduction sur papier ou sur format électronique. The author retains ownership of the L'auteur conserve la propriété du copyright in this thesis. Neither the droit d'auteur qui protège cette thèse. thesis nor substantial extracts fkom it Ni la thèse ni des extraits substantiels may be printed or otherwise de celle-ci ne doivent être imprimés reproduced without the author's ou autrement reproduits sans son permission. autorisation. Functional Analysis of The Homeobox Gene TLr-2 During Mouse Embryogenesis Doctor of Philosophy (1998) Shao Jun Tang Graduate Department of Moiecular and Medicd Genetics University of Toronto Abstract This thesis describes the clonhg of the TLx-2 homeobox gene, the determination of its developmental expression, the characterization of its fiuiction in mouse mesodem and penpheral nervous system (PNS) developrnent, the regulation of nx-2 expression in the early mouse embryo by BMP signalling, and the modulation of the function of nX-2 protein by the 14-3-3 signalling protein during neural development. -

Nhbs Annual New and Forthcoming Titles Issue: 2000 Complete January 2001 [email protected] +44 (0)1803 865913
nhbs annual new and forthcoming titles Issue: 2000 complete January 2001 [email protected] +44 (0)1803 865913 The NHBS Monthly Catalogue in a complete yearly edition Zoology: Mammals Birds Welcome to the Complete 2000 edition of the NHBS Monthly Catalogue, the ultimate Reptiles & Amphibians buyer's guide to new and forthcoming titles in natural history, conservation and the Fishes environment. With 300-400 new titles sourced every month from publishers and research organisations around the world, the catalogue provides key bibliographic data Invertebrates plus convenient hyperlinks to more complete information and nhbs.com online Palaeontology shopping - an invaluable resource. Each month's catalogue is sent out as an HTML Marine & Freshwater Biology email to registered subscribers (a plain text version is available on request). It is also General Natural History available online, and offered as a PDF download. Regional & Travel Please see our info page for more details, also our standard terms and conditions. Botany & Plant Science Prices are correct at the time of publication, please check www.nhbs.com for the Animal & General Biology latest prices. Evolutionary Biology Ecology Habitats & Ecosystems Conservation & Biodiversity Environmental Science Physical Sciences Sustainable Development Data Analysis Reference Mammals Activity Patterns in Small Mammals 318 pages | 59 figs, 11 tabs | Springer An Ecological Approach Hbk | 2000 | 354059244X | #109391A | Edited by S Halle and NC Stenseth £100.00 BUY Links chronobiology with behavioural and evolutionary ecology, drawing on research on mammals ranging from mongooses and civets to weasels, martens and shrews. .... African Rhino 92 pages | B/w photos, figs, tabs | IUCN Status Survey and Conservation Action Plan Pbk | 1999 | 2831705029 | #106031A | Richard Emslie and Martin Brooks £15.00 BUY Action plan aimed at donors, government and non-government organisations, and all those involved in rhino conservation. -

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -
BSCB Newsletter 2017D
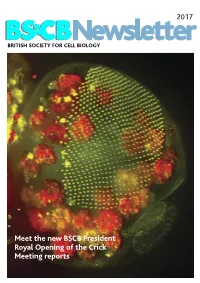
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

July/August 2011
ASPB News THE NEWSLETTER OF THE AMERICAN SOCIETY OF PLANT BIOLOGISTS Volume 38, Number 4 July/August 2011 Inside This Issue ASPB Members Elected to 2011 Class President’s Letter of National Academy of Sciences 75th Annual Meeting of the Northeastern Section ASPB members Steven E. Jacobsen and James A. recognition of their distinguished and continuing Birchler were elected to the National Academy of Sci- achievements in original research. The total number ASPB Welcomes Patti ences (NAS) on May 3, 2011, at the Academy’s 148th of active members is now 2,113, and the total Lockhart as New Managing Editor annual meeting. In addition, ASPB member Jiayang number of foreign associates is 418. Foreign associ- Li was named as a foreign associate of the Academy. ates are nonvoting members of the Academy, with Kathy Munkvold Selected This election admitted a total of 72 new mem- citizenship outside the United States. as ASPB Plant Science Policy Fellow bers and 18 foreign associates from 15 countries in continued on page 13 HHMI and GBMF Name 15 ASPB Members as Investigators Two of the nation’s largest private sponsors of in fundamental plant science, and we hope it will research have taken a giant leap into plant science. encourage others in the United States to make The Howard Hughes Medical Institute (HHMI) and analogous commitments.” the Gordon and Betty Moore Foundation (GBMF) Vicki L. Chandler, a former ASPB president who have named 15 of the country’s most innovative is GBMF chief program officer for science, said that plant scientists as HHMI-GBMF Investigators. -

Female Fellows of the Royal Society
Female Fellows of the Royal Society Professor Jan Anderson FRS [1996] Professor Ruth Lynden-Bell FRS [2006] Professor Judith Armitage FRS [2013] Dr Mary Lyon FRS [1973] Professor Frances Ashcroft FMedSci FRS [1999] Professor Georgina Mace CBE FRS [2002] Professor Gillian Bates FMedSci FRS [2007] Professor Trudy Mackay FRS [2006] Professor Jean Beggs CBE FRS [1998] Professor Enid MacRobbie FRS [1991] Dame Jocelyn Bell Burnell DBE FRS [2003] Dr Philippa Marrack FMedSci FRS [1997] Dame Valerie Beral DBE FMedSci FRS [2006] Professor Dusa McDuff FRS [1994] Dr Mariann Bienz FMedSci FRS [2003] Professor Angela McLean FRS [2009] Professor Elizabeth Blackburn AC FRS [1992] Professor Anne Mills FMedSci FRS [2013] Professor Andrea Brand FMedSci FRS [2010] Professor Brenda Milner CC FRS [1979] Professor Eleanor Burbidge FRS [1964] Dr Anne O'Garra FMedSci FRS [2008] Professor Eleanor Campbell FRS [2010] Dame Bridget Ogilvie AC DBE FMedSci FRS [2003] Professor Doreen Cantrell FMedSci FRS [2011] Baroness Onora O'Neill * CBE FBA FMedSci FRS [2007] Professor Lorna Casselton CBE FRS [1999] Dame Linda Partridge DBE FMedSci FRS [1996] Professor Deborah Charlesworth FRS [2005] Dr Barbara Pearse FRS [1988] Professor Jennifer Clack FRS [2009] Professor Fiona Powrie FRS [2011] Professor Nicola Clayton FRS [2010] Professor Susan Rees FRS [2002] Professor Suzanne Cory AC FRS [1992] Professor Daniela Rhodes FRS [2007] Dame Kay Davies DBE FMedSci FRS [2003] Professor Elizabeth Robertson FRS [2003] Professor Caroline Dean OBE FRS [2004] Dame Carol Robinson DBE FMedSci -

The Control of Shoot Branching: an Example of Plant Information Processing
Plant, Cell and Environment (2009) 32, 694–703 doi: 10.1111/j.1365-3040.2009.01930.x The control of shoot branching: an example of plant information processing OTTOLINE LEYSER Department of Biology, Area 11, University of York, York YO10 5YW, UK ABSTRACT the apical–basal axis defined by the establishment of the shoot apical meristem at one end, and the root apical mer- Throughout their life cycle, plants adjust their body plan istem at the other. Post-embryonically, the meristems give to suit the environmental conditions in which they are rise to the entire shoot and root systems, respectively. Fur- growing. A good example of this is in the regulation of thermore, the tissues they establish can produce secondary shoot branching. Axillary meristems laid down in each leaf meristems, which if activated can produce entirely new formed from the primary shoot apical meristem can remain axes of growth with the same developmental potential as dormant, or activate to produce a branch. The decision the primary root or shoot from which they were derived. whether to activate an axillary meristem involves the assess- Thus, the body plan of a plant is determined continuously ment of a wide range of external environmental, internal throughout its life cycle, allowing it to be exquisitely envi- physiological and developmental factors. Much of this infor- ronmentally responsive. Plants can alter their body plan to mation is conveyed to the axillary meristem via a network suit their environment, and thus the environmentally regu- of interacting hormonal signals that can integrate inputs lated development of new growth axes is functionally from diverse sources, combining multiple local signals to equivalent to environmentally regulated animal behav- generate a rich source of systemically transmitted informa- iours. -

Helping Diabetes Sufierers
Helping diabetes sufferers Thanks to work carried out by researchers at the University of Oxford, patients with a particular genetic form of diabetes can now be treated with an oral tablet rather than an insulin injection, which dramatically improves their blood glucose control and quality of life. www.ox.ac.uk/oxfordimpacts In the past, people with neonatal diabetes faced a life of insulin injections. However, the discovery that their diabetes is due to activating mutations in the potassium channel has enabled them to switch to sulphonylurea tablet therapy. This has not only enhanced their blood sugar control and lowered their risk of developing the secondary complications of diabetes; it has also improved their quality of life and that of their families. In a few rare cases, patients with specific genetic defects not only have diabetes, they also show developmental delay, and start to walk and talk much later than expected for their age. Treatment with sulphonylurea tablets can often help these problems too. These patients have neonatal diabetes, a rare Professors Ashcroft and Hattersley organised the type of diabetes that usually develops within six first meeting for patients with neonatal diabetes months of birth. It results from a genetic defect and their families in July 2009, which enabled in a protein known as a potassium channel, which them to meet one another as well as the scientists is crucially important for ensuring that insulin is involved in the research. secreted when blood sugar levels rise. Professor Frances Ashcroft from the Department of Physiology, Anatomy and Genetics was the first to discover the potassium channel and to show how it controls insulin secretion. -

Sulfonylurea Stimulation of Insulin Secretion Peter Proks,1 Frank Reimann,2 Nick Green,1 Fiona Gribble,2 and Frances Ashcroft1
Sulfonylurea Stimulation of Insulin Secretion Peter Proks,1 Frank Reimann,2 Nick Green,1 Fiona Gribble,2 and Frances Ashcroft1 Sulfonylureas are widely used to treat type 2 diabetes smooth, and skeletal muscle, and some brain neurones. In because they stimulate insulin secretion from pancre- all these tissues, opening of KATP channels in response to atic -cells. They primarily act by binding to the SUR metabolic stress leads to inhibition of electrical activity. subunit of the ATP-sensitive potassium (KATP) channel Thus they are involved in the response to both cardiac and and inducing channel closure. However, the channel is cerebral ischemia (2). They are also important in neuronal still able to open to a limited extent when the drug is regulation of glucose homeostasis (3), seizure protection bound, so that high-affinity sulfonylurea inhibition is not complete, even at saturating drug concentrations. (4), and the control of vascular smooth muscle tone (and, thereby, blood pressure) (5). KATP channels are also found in cardiac, skeletal, and smooth muscle, but in these tissues are composed of The KATP channel is a hetero-octameric complex of two different SUR subunits that confer different drug different types of protein subunits: an inwardly rectifying sensitivities. Thus tolbutamide and gliclazide block Kϩ channel, Kir6.x, and a sulfonylurea receptor, SUR (6,7).  ϩ channels containing SUR1 ( -cell type), but not SUR2 Kir6.x belongs to the family of inwardly rectifying K (cardiac, smooth muscle types), whereas glibenclamide, (Kir) channels and assembles as a tetramer to form the glimepiride, repaglinide, and meglitinide block both types of channels. -

Alternative Splicing Modulates Ubx Protein Function in Drosophila Melanogaster
Copyright Ó 2010 by the Genetics Society of America DOI: 10.1534/genetics.109.112086 Alternative Splicing Modulates Ubx Protein Function in Drosophila melanogaster Hilary C. Reed,* Tim Hoare,† Stefan Thomsen,‡ Thomas A. Weaver,† Robert A. H. White,† Michael Akam* and Claudio R. Alonso‡,1 *Department of Zoology, University of Cambridge, Cambridge CB2 3EJ, United Kingdom, †Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge CB2 3DY, United Kingdom and ‡School of Life Sciences, University of Sussex, Brighton BN1 9QG, United Kingdom Manuscript received November 13, 2009 Accepted for publication December 17, 2009 ABSTRACT The Drosophila Hox gene Ultrabithorax (Ubx) produces a family of protein isoforms through alternative splicing. Isoforms differ from one another by the presence of optional segments—encoded by individual exons—that modify the distance between the homeodomain and a cofactor-interaction module termed the ‘‘YPWM’’ motif. To investigate the functional implications of Ubx alternative splicing, here we analyze the in vivo effects of the individual Ubx isoforms on the activation of a natural Ubx molecular target, the decapentaplegic (dpp) gene, within the embryonic mesoderm. These experiments show that the Ubx isoforms differ in their abilities to activate dpp in mesodermal tissues during embryogenesis. Furthermore, using a Ubx mutant that reduces the full Ubx protein repertoire to just one single isoform, we obtain specific anomalies affecting the patterning of anterior abdominal muscles, demonstrating that Ubx isoforms are not functionally interchangeable during embryonic mesoderm development. Finally, a series of experiments in vitro reveals that Ubx isoforms also vary in their capacity to bind DNA in presence of the cofactor Extradenticle (Exd). -

HOX CLUSTER INTERGENIC SEQUENCE EVOLUTION by JEREMY DON RAINCROW a Dissertation Submitted to the Graduate School-New Brunswick R
HOX CLUSTER INTERGENIC SEQUENCE EVOLUTION By JEREMY DON RAINCROW A Dissertation submitted to the Graduate School-New Brunswick Rutgers, The State University of New Jersey and The Graduate School of Biomedical Sciences University of Medicine and Dentistry of New Jersey in partial fulfillment of the requirements for the degree of Doctor of Philosophy Graduate Program in Cell and Developmental Biology written under the direction of Chi-hua Chiu and approved by ______________________________________________ ______________________________________________ ______________________________________________ ______________________________________________ New Brunswick, New Jersey October 2010 ABSTRACT OF THE DISSERTATION HOX GENE CLUSTER INTERGENIC SEQUENCE EVOLUTION by JEREMY DON RAINCROW Dissertation Director: Chi-hua Chiu The Hox gene cluster system is highly conserved among jawed-vertebrates. Specifically, the coding region of Hox genes along with their spacing and occurrence is highly conserved throughout gnathostomes. The intergenic regions of these clusters however are more variable. During the construction of a comprehensive non-coding sequence database we discovered that the intergenic sequences appear to also be highly conserved among cartilaginous and lobe-finned fishes, but much more diverged and dynamic in the ray-finned fishes. Starting at the base of the Actinopterygii a turnover of otherwise highly conserved non-coding sequences begins. This turnover is extended well into the derived ray-finned fish clade, Teleostei. Evidence from our population genetic study suggests this turnover, which appears to be due mainly to loosened constraints at the macro-evolutionary level, is highlighted by evidence of strong positive selection acting at the micro-evolutionary level. During the construction of the non-coding sequence database we also discovered that along with evidence of both relaxed constraints and positive selection emerges a pattern of transposable elements found within the Hox gene cluster system. -

Homeotic Gene Action in Embryonic Brain Development of Drosophila
Development 125, 1579-1589 (1998) 1579 Printed in Great Britain © The Company of Biologists Limited 1998 DEV1254 Homeotic gene action in embryonic brain development of Drosophila Frank Hirth, Beate Hartmann and Heinrich Reichert* Institute of Zoology, University of Basel, Rheinsprung 9, CH-4051 Basel, Switzerland *Author for correspondence (e-mail: [email protected]) Accepted 18 February; published on WWW 1 April 1998 SUMMARY Studies in vertebrates show that homeotic genes are absence of labial, mutant cells are generated and positioned involved in axial patterning and in specifying segmental correctly in the brain, but these cells do not extend axons. identity of the embryonic hindbrain and spinal cord. To Additionally, extending axons of neighboring wild-type gain further insights into homeotic gene action during CNS neurons stop at the mutant domains or project ectopically, development, we here characterize the role of the homeotic and defective commissural and longitudinal pathways genes in embryonic brain development of Drosophila. We result. Immunocytochemical analysis demonstrates that first use neuroanatomical techniques to map the entire cells in the mutant domains do not express neuronal anteroposterior order of homeotic gene expression in the markers, indicating a complete lack of neuronal identity. Drosophila CNS, and demonstrate that this order is An alternative glial identity is not adopted by these mutant virtually identical in the CNS of Drosophila and mammals. cells. Comparable effects are seen in Deformed mutants but We then carry out a genetic analysis of the labial gene in not in other homeotic gene mutants. Our findings embryonic brain development. Our analysis shows that demonstrate that the action of the homeotic genes labial loss-of-function mutation and ubiquitous overexpression of and Deformed are required for neuronal differentiation in labial results in ectopic expression of neighboring the developing brain of Drosophila.