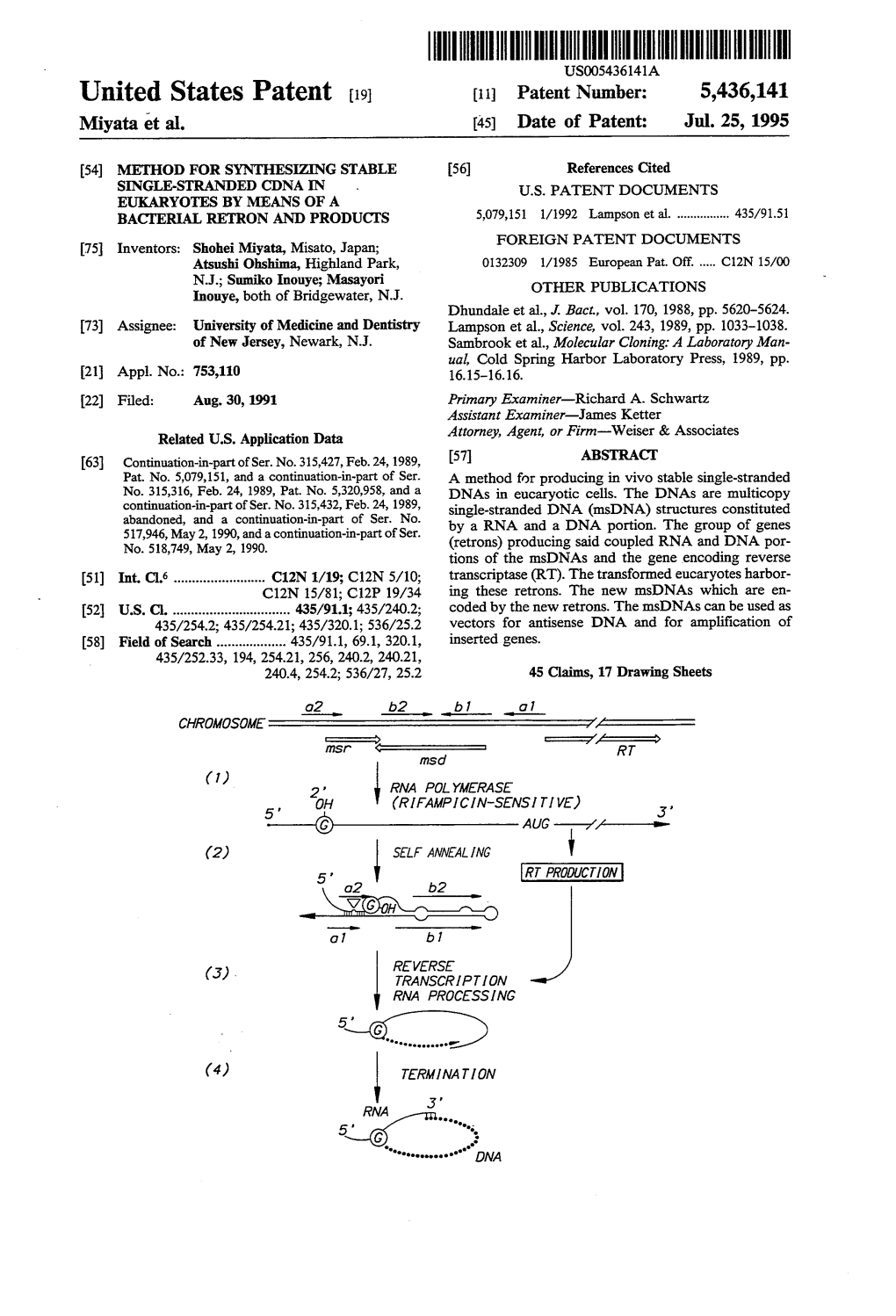
Neo-Cloo2 B2 O 1 B 1 (3) REVERSE TRANSCRIPTION RNA PROCESSING
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group. -

Mirna Detection Using a Rolling Circle Amplification and RNA
biosensors Article MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy Chenxin Fang, Ping Ouyang, Yuxing Yang, Yang Qing, Jialun Han, Wenyan Shang, Yubing Chen and Jie Du * State Key Laboratory of Marine Resource Utilization in South China Sea, College of Materials Science and Engineering, Hainan University, Haikou 570228, China; [email protected] (C.F.); [email protected] (P.O.); [email protected] (Y.Y.); [email protected] (Y.Q.); [email protected] (J.H.); [email protected] (W.S.); [email protected] (Y.C.) * Correspondence: [email protected] Abstract: A microRNA (miRNA) detection platform composed of a rolling circle amplification (RCA) system and an allosteric deoxyribozyme system is proposed, which can detect miRNA-21 rapidly and efficiently. Padlock probe hybridization with the target miRNA is achieved through complementary base pairing and the padlock probe forms a closed circular template under the action of ligase; this circular template results in RCA. In the presence of DNA polymerase, RCA proceeds and a long chain with numerous repeating units is formed. In the presence of single-stranded DNA (H1 and H2), multi-component nucleic acid enzymes (MNAzymes) are formed that have the ability to cleave substrates. Finally, substrates containing fluorescent and quenching groups and magnesium ions are added to the system to activate the MNAzyme and the substrate cleavage reaction, thus achieving fluorescence intensity amplification. The RCA–MNAzyme system has dual signal amplification and presents a sensing platform that demonstrates broad prospects in the analysis and detection of Citation: Fang, C.; Ouyang, P.; Yang, nucleic acids. -

De Novo Nucleic Acids: a Review of Synthetic Alternatives to DNA and RNA That Could Act As † Bio-Information Storage Molecules
life Review De Novo Nucleic Acids: A Review of Synthetic Alternatives to DNA and RNA That Could Act as y Bio-Information Storage Molecules Kevin G Devine 1 and Sohan Jheeta 2,* 1 School of Human Sciences, London Metropolitan University, 166-220 Holloway Rd, London N7 8BD, UK; [email protected] 2 Network of Researchers on the Chemical Evolution of Life (NoR CEL), Leeds LS7 3RB, UK * Correspondence: [email protected] This paper is dedicated to Professor Colin B Reese, Daniell Professor of Chemistry, Kings College London, y on the occasion of his 90th Birthday. Received: 17 November 2020; Accepted: 9 December 2020; Published: 11 December 2020 Abstract: Modern terran life uses several essential biopolymers like nucleic acids, proteins and polysaccharides. The nucleic acids, DNA and RNA are arguably life’s most important, acting as the stores and translators of genetic information contained in their base sequences, which ultimately manifest themselves in the amino acid sequences of proteins. But just what is it about their structures; an aromatic heterocyclic base appended to a (five-atom ring) sugar-phosphate backbone that enables them to carry out these functions with such high fidelity? In the past three decades, leading chemists have created in their laboratories synthetic analogues of nucleic acids which differ from their natural counterparts in three key areas as follows: (a) replacement of the phosphate moiety with an uncharged analogue, (b) replacement of the pentose sugars ribose and deoxyribose with alternative acyclic, pentose and hexose derivatives and, finally, (c) replacement of the two heterocyclic base pairs adenine/thymine and guanine/cytosine with non-standard analogues that obey the Watson–Crick pairing rules. -

Design and Switch of Catalytic Activity with the Dnazyme–Rnazyme Combination
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 581 (2007) 1763–1768 Design and switch of catalytic activity with the DNAzyme–RNAzyme combination Yongjie Sheng, Zhen Zeng, Wei Peng, Dazhi Jiang, Shuang Li, Yanhong Sun, Jin Zhang* Key Laboratory for Molecular Enzymology and Engineering of Ministry of Education, Jilin University, Changchun 130023, PR China Received 12 January 2007; revised 9 March 2007; accepted 16 March 2007 Available online 2 April 2007 Edited by Judit Ova´di backbone DNA with a regulating sequence [12]. Compared to Abstract Design and switch of catalytic activity in enzymology remains a subject of intense investigation. Here, we employ a the linear hammerhead ribozyme, the stability of the circular DNAzyme–RNAzyme combination strategy for construction of RNA–DNA enzyme was substantially enhanced. Furthering a 10–23 deoxyribozyme-hammerhead ribozyme combination that this approach, we substituted the hammerhead ribozyme with targets different sites of the b-lactamase mRNA. The 10–23 the more efficient and more stable 10–23 deoxyribozyme, while deoxyribozyme-hammerhead ribozyme combination gene was the backbone DNA was replaced by the single-stranded repli- cloned into phagemid vector pBlue-scriptIIKS (+). In vitro the cation-competent vector M13mp18 (7.25 kb), thus successfully single-strand recombinant phagemid vector containing the com- constructing a novel replicating circular deoxyribozyme, which bination sequence exhibited 10–23 deoxyribozyme activity, and displayed 10–23 deoxyribozyme activities both in vitro and in the linear transcript displayed hammerhead ribozyme activity. bacteria [13,14]. In bacteria, the 10–23 deoxyribozyme-hammerhead ribozyme Here, we report the combination of 10–23 deoxyribozyme combination inhibited the b-lactamase expression and repressed the growth of drug-resistant bacteria. -

LESSON 4 Using Bioinformatics to Analyze Protein Sequences
LESSON 4 Using Bioinformatics to 4 Analyze Protein Sequences Introduction In this lesson, students perform a paper exercise designed to reinforce the student understanding of the complementary nature of DNA and how that complementarity leads to six potential protein reading frames in any given DNA sequence. They also gain familiarity with the circular format codon table. Students then use the bioinformatics tool ORF Finder to identify the reading frames in their DNA sequence from Lesson Two and Lesson Three, and to select Class Time the proper open reading frame to use in a multiple sequence alignment with 2 class periods (approximately 50 their protein sequences. In Lesson Four, students also learn how biological minutes each). anthropologists might use bioinformatics tools in their career. Prior Knowledge Needed • DNA contains the genetic information Learning Objectives that encodes traits. • DNA is double stranded and At the end of this lesson, students will know that: anti-parallel. • Each DNA molecule is composed of two complementary strands, which are • The beginning of a DNA strand is arranged anti-parallel to one another. called the 5’ (“five prime”) region and • There are three potential reading frames on each strand of DNA, and a total the end of a DNA strand is called the of six potential reading frames for protein translation in any given region of 3’ (“three prime”) region. the DNA molecule (three on each strand). • Proteins are produced through the processes of transcription and At the end of this lesson, students will be able to: translation. • Amino acids are encoded by • Identify the best open reading frame among the six possible reading frames nucleotide triplets called codons. -

Conditional Deoxyribozyme−Nanoparticle Conjugates For
www.acsami.org Research Article Conditional Deoxyribozyme−Nanoparticle Conjugates for miRNA- Triggered Gene Regulation Jiahui Zhang, Rong Ma, Aaron Blanchard, Jessica Petree, Hanjoong Jo,* and Khalid Salaita* Cite This: ACS Appl. Mater. Interfaces 2020, 12, 37851−37861 Read Online ACCESS Metrics & More Article Recommendations *sı Supporting Information ABSTRACT: DNA−nanoparticle (NP) conjugates have been used to knockdown gene expression transiently and effectively, making them desirable tools for gene regulation therapy. Because DNA−NPs are constitutively active and are rapidly taken up by most cell types, they offer limited control in terms of tissue or cell type specificity. To take a step toward solving this issue, we incorporate toehold-mediated strand exchange, a versatile molec- ular programming modality, to switch the DNA−NPs from an inactive state to an active state in the presence of a specific RNA input. Because many transcripts are unique to cell subtype or disease state, this approach could one day lead to responsive nucleic acid therapeutics with enhanced specificity. As a proof of concept, we designed conditional deoxyribozyme−nanoparticles (conditional DzNPs) that knockdown tumor necrosis factor α (TNFα) mRNA upon miR-33 triggering. We demonstrate toehold- mediated strand exchange and restoration of TNFα DNAzyme activity in the presence of miR-33 trigger, with optimization of the preparation, configuration, and toehold length of conditional DzNPs. Our results indicate specific and strong ON/OFF response of conditional DzNPs to the miR-33 trigger in buffer. Furthermore, we demonstrate endogenous miR-33-triggered knockdown of TNFα mRNA in mouse macrophages, implying the potential of conditional gene regulation applications using these DzNPs. -

Cytoplasmic Synthesis of Endogenous Alu Complementary DNA Via Reverse Transcription and Implications in Age-Related Macular Degeneration
Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration Shinichi Fukudaa,b,c,1, Akhil Varshneya,b,1, Benjamin J. Fowlerd, Shao-bin Wanga,b, Siddharth Narendrana,b,e, Kameshwari Ambatia,b, Tetsuhiro Yasumad,f, Joseph Magagnolig,h, Hannah Leunga,b, Shuichiro Hiraharaa,b,i, Yosuke Nagasakaa,b, Reo Yasumaa,b,f, Ivana Apicellaa,b, Felipe Pereiraa,b,j, Ryan D. Makina,b, Eamonn Magnerk, Xinan Liuk, Jian Suna,b, Mo Wangl, Kirstie Bakerl, Kenneth M. Marionl, Xiwen Huangk, Elmira Baghdasaryanl,m, Meenakshi Ambatia,b,n, Vidya L. Ambatin, Akshat Pandeya,b, Lekha Pandyaa,b, Tammy Cummingsg,h, Daipayan Banerjeea,b, Peirong Huanga,b, Praveen Yerramothua,b, Genrich V. Tolstonogo, Ulrike Heldp, Jennifer A. Erwinq, Apua C. M. Paquolaq, Joseph R. Herdyr, Yuichiro Ogurai, Hiroko Terasakif, Tetsuro Oshikac, Shaban Darwishs,t, Ramendra K. Singhs, Saghar Mozaffaris, Deepak Bhattaraiu, Kyung Bo Kimu, James W. Harding,v, Charles L. Bennettg,h,w, David R. Hintonx,y, Timothy E. Hansonz,aa, Christian Röverbb, Keykavous Parangs, Nagaraj Kerura,b,cc,dd, Jinze Liuk, Brian C. Werneree, S. Scott Suttong,h, Srinivas R. Saddal,m, Gerald G. Schumannp, Bradley D. Gelfanda,b,ff, Fred H. Gager,2, and Jayakrishna Ambatia,b,dd,gg,2 aCenter for Advanced Vision Science, School of Medicine, University of Virginia, Charlottesville, VA 22908; bDepartment of Ophthalmology, School of Medicine, University of Virginia, Charlottesville, VA 22908; cDepartment of Ophthalmology, University of Tsukuba, Ibaraki -

Transitioning to DNA Genomes in an RNA World
INSIGHT ORIGIN OF LIFE Transitioning to DNA genomes in an RNA world The unexpected ability of an RNA polymerase ribozyme to copy RNA into DNA has ramifications for understanding how DNA genomes evolved. RAZVAN COJOCARU AND PETER J UNRAU In modern metabolism, protein-based Related research article Samanta B, Joyce enzymes called reverse transcriptases can copy GF. 2017. A reverse transcriptase ribozyme. RNA to produce molecules of complementary eLife 6:e31153. DOI: 10.7554/eLife.31153 DNA. Other enzymes can promote the produc- tion of DNA nucleotides (the building blocks of DNA molecules) from RNA nucleotides via chal- lenging chemical reactions. So how did the first DNA genomes come to be? There are two pos- or as long as history has been recorded, sibilities within the framework of the RNA world. humanity has tried to answer the ancient In the first, protein enzymes evolved before question of our origins. The ‘central DNA genomes. In the second, the RNA world F contained RNA polymerase ribozymes that were dogma’ of molecular biology, first stated by Fran- able to produce single-stranded complementary cis Crick in 1958, represented a major step for- DNA and then convert it into stable double- ward in our efforts to answer this question stranded DNA genomes. (Figure 1A; Crick, 1958). In this model, the A number of laboratories around the world genetic information stored in DNA is transcribed are trying to build ribozymes that can sustain to produce RNA, which is then translated by the RNA replication (Wang et al., 2011; ribosome to produce chains of amino acids. -

Plasmid Rolling Circle Replication: Identification of the RNA
The EMBO Journal Vol.16 No.18 pp.5784–5795, 1997 Plasmid rolling circle replication: identification of the RNA polymerase-directed primer RNA and requirement for DNA polymerase I for lagging strand synthesis M.Gabriela Kramer, Saleem A.Khan1 and (del Solar et al., 1987a; Gruss et al., 1987). It has been Manuel Espinosa2 postulated that lagging strand synthesis is initiated by a primer RNA (pRNA) which is synthesized from the sso Centro de Investigaciones Biolo´gicas, CSIC, Velazquez, 144, (Birch and Khan, 1992; Dempsey et al., 1995). Primer 1 E-28006 Madrid, Spain and Department of Molecular Genetics and synthesis is thought to be performed by the host RNA Biochemistry, University of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA polymerase (RNAP) since the presence of the RNAP inhibitor rifampicin leads to: (i) in vivo accumulation of 2Corresponding author e-mail: [email protected] ssDNA (Boe et al., 1989; Kramer et al., 1995); (ii) inhibition of in vitro DNA replication of the RCR plasmid Plasmid rolling circle replication involves generation pLS1 (del Solar et al., 1987b); and (iii) inhibition of in of single-stranded DNA (ssDNA) intermediates. ssDNA vitro DNA synthesis from ssDNA templates prepared from released after leading strand synthesis is converted to various RCR plasmids (Birch and Khan, 1992; Dempsey a double-stranded form using solely host proteins. et al., 1995). However, based on either sequence homo- Most plasmids that replicate by the rolling circle mode logies or partial inhibition by rifampicin, it has been contain palindromic sequences that act as the single proposed that lagging strand replication may also be strand origin, sso.