De Novo Nucleic Acids: a Review of Synthetic Alternatives to DNA and RNA That Could Act As † Bio-Information Storage Molecules
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Addressing Evolutionary Questions with Synthetic Biology
Addressing evolutionary questions with synthetic biology Florian Baier and Yolanda Schaerli Department of Fundamental Microbiology, University of Lausanne, Biophore Building, 1015 Lausanne, Switzerland Correspondence: [email protected]; [email protected] Abstract Synthetic biology emerged as an engineering discipline to design and construct artificial biological systems. Synthetic biological designs aim to achieve specific biological behavior, which can be exploited for biotechnological, medical and industrial purposes. In addition, mimicking natural systems using well-characterized biological parts also provides powerful experimental systems to study evolution at the molecular and systems level. A strength of synthetic biology is to go beyond nature’s toolkit, to test alternative versions and to study a particular biological system and its phenotype in isolation and in a quantitative manner. Here, we review recent work that implemented synthetic systems, ranging from simple regulatory circuits, rewired cellular networks to artificial genomes and viruses, to study fundamental evolutionary concepts. In particular, engineering, perturbing or subjecting these synthetic systems to experimental laboratory evolution provides a mechanistic understanding on important evolutionary questions, such as: Why did particular regulatory networks topologies evolve and not others? What happens if we rewire regulatory networks? Could an expanded genetic code provide an evolutionary advantage? How important is the structure of genome and number of chromosomes? Although the field of evolutionary synthetic biology is still in its teens, further advances in synthetic biology provide exciting technologies and novel systems that promise to yield fundamental insights into evolutionary principles in the near future. 1 1. Introduction Evolutionary biology traditionally studies past or present organisms to reconstruct past evolutionary events with the aim to explain and predict their evolution. -

Significance and Implications of Vitamin B-12 Reaction Shema- ETH ZURICH VARIANT: Mechanisms and Insights
Taylor University Pillars at Taylor University Student Scholarship: Chemistry Chemistry and Biochemistry Fall 2019 Significance and Implications of Vitamin B-12 Reaction Shema- ETH ZURICH VARIANT: Mechanisms and Insights David Joshua Ferguson Follow this and additional works at: https://pillars.taylor.edu/chemistry-student Part of the Analytical Chemistry Commons, Inorganic Chemistry Commons, Organic Chemistry Commons, Other Chemistry Commons, and the Physical Chemistry Commons CHEMISTRY THESIS SIGNIFICANCE AND IMPLICATIONS OF VITAMIN B-12 REACTION SCHEMA- ETH ZURICH VARIANT: MECHANISMS AND INSIGHTS DAVID JOSHUA FERGUSON 2019 2 Table of Contents: Chapter 1 6 Chapter 2 17 Chapter 3 40 Chapter 4 59 Chapter 5 82 Chapter 6 118 Chapter 7 122 Appendix References 3 Chapter 1 A. INTRODUCTION. Vitamin B-12 otherwise known as cyanocobalamin is a compound with synthetic elegance. Considering how it is composed of an aromatic macrocyclic corrin there are key features of this molecule that are observed either in its synthesis of in the biochemical reactions it plays a role in whether they be isomerization reactions or transfer reactions. In this paper the focus for the discussion will be on the history, chemical significance and total synthesis of vitamin B12. Even more so the paper will be concentrated one of the two variants of the vitamin B-12 synthesis, namely the ETH Zurich variant spearheaded by Albert Eschenmoser.Examining the structure as a whole it is observed that a large portion of the vitamin B12 is a corrin structure with a cobalt ion in the center of the macrocyclic part, and that same cobalt ion has cyanide ligands. -

Rnase a Cleanup of DNA Samples Version Number: 1.0 Version 1.0 Date: 12/21/2016 Author(S): Yuko Yoshinaga, Eileen Dalin Reviewed/Revised By
SAMPLE MANAGEMENT STANDARD OPERATING PROCEDURE RNase A Cleanup of DNA Samples Version Number: 1.0 Version 1.0 Date: 12/21/2016 Author(s): Yuko Yoshinaga, Eileen Dalin Reviewed/Revised by: Summary RNase A treatment is used for the removal of RNA from genomic DNA samples. RNase A cleaves the phosphodiester bond between the 5'-ribose of a nucleotide and the phosphate group attached to the 3'- ribose of an adjacent pyrimidine nucleotide. The resulting 2', 3'-cyclic phosphate is hydrolyzed to the corresponding 3'-nucleoside phosphate. Materials & Reagents Materials Vendor Stock Number Disposables 1.5 ml microcentrifuge tubes Reagents TE Buffer, pH 8.0 Ambion 9849 RNase A, DNase and protease-free (10 mg/ml) ThermoFisher EN0531 Sodium acetate buffer solution (3M, pH 5.2) VWR 567422 Ethanol, 200 proof Pharmco-Aaper 111000200CSPP 50x TAE Buffer Life Technologies/ Invitrogen MRGF-4210 SYBR® Safe DNA gel stain (10,000x concentrate in Life Technologies/ Invitrogen S33102 DMSO) DNA Molecular Weight Marker II Sigma 10236250001 Gel Loading Dye, Blue (6x) New England BioLabs B7021S Equipment Centrifuge 4°C Heat block 37°C Gel electrophoresis device Gel Imager Bio-Rad EH&S PPE Requirements: 1.1 Safety glasses, lab coat, and nitrile gloves should be worn at all times while performing work in the lab during this protocol. 1.2 Ethanol is a highly flammable and irritating to the eyes. Vapors may cause drowsiness and dizziness. Keep containers closed and keep away from sources of ignition such as smoking. 1 SAMPLE MANAGEMENT STANDARD OPERATING PROCEDURE Avoid contact with skin and eyes. In case of contact with eyes, rinse immediately with plenty of water and seek medical advice. -

DNA Microarrays (Gene Chips) and Cancer
DNA Microarrays (Gene Chips) and Cancer Cancer Education Project University of Rochester DNA Microarrays (Gene Chips) and Cancer http://www.biosci.utexas.edu/graduate/plantbio/images/spot/microarray.jpg http://www.affymetrix.com Part 1 Gene Expression and Cancer Nucleus Proteins DNA RNA Cell membrane All your cells have the same DNA Sperm Embryo Egg Fertilized Egg - Zygote How do cells that have the same DNA (genes) end up having different structures and functions? DNA in the nucleus Genes Different genes are turned on in different cells. DIFFERENTIAL GENE EXPRESSION GENE EXPRESSION (Genes are “on”) Transcription Translation DNA mRNA protein cell structure (Gene) and function Converts the DNA (gene) code into cell structure and function Differential Gene Expression Different genes Different genes are turned on in different cells make different mRNA’s Differential Gene Expression Different genes are turned Different genes Different mRNA’s on in different cells make different mRNA’s make different Proteins An example of differential gene expression White blood cell Stem Cell Platelet Red blood cell Bone marrow stem cells differentiate into specialized blood cells because different genes are expressed during development. Normal Differential Gene Expression Genes mRNA mRNA Expression of different genes results in the cell developing into a red blood cell or a white blood cell Cancer and Differential Gene Expression mRNA Genes But some times….. Mutations can lead to CANCER CELL some genes being Abnormal gene expression more or less may result -

Dna the Code of Life Worksheet
Dna The Code Of Life Worksheet blinds.Forrest Jowled titter well Giffy as misrepresentsrecapitulatory Hughvery nomadically rubberized herwhile isodomum Leonerd exhumedremains leftist forbiddenly. and sketchable. Everett clem invincibly if arithmetical Dawson reinterrogated or Rewriting the Code of Life holding for Genetics and Society. C A process look a genetic code found in DNA is copied and converted into value chain of. They may negatively impact of dna worksheet answers when published by other. Cracking the Code of saw The Biotechnology Institute. DNA lesson plans mRNA tRNA labs mutation activities protein synthesis worksheets and biotechnology experiments for open school property school biology. DNA the code for life FutureLearn. Cracked the genetic code to DNA cloning twins and Dolly the sheep. Dna are being turned into consideration the code life? DNA The Master Molecule of Life CDN. This window or use when he has been copied to a substantial role in a qualified healthcare professional journals as dna the pace that the class before scientists have learned. Explore the Human Genome Project within us Learn about DNA and genomics role in medicine and excellent at the Smithsonian National Museum of Natural. DNA The Double Helix. Most enzymes create a dna the code of life worksheet is getting the. Worksheet that describes the structure of DNA students color the model according to instructions Includes a. Biology Materials Handout MA-H2 Microarray Virtual Lab Activity Worksheet. This user has, worksheet the dna code of life, which proteins are carried on. Notes that scientists have worked 10 years to disappoint the manner human genome explains that DNA is a chemical message that began more data four billion years ago. -
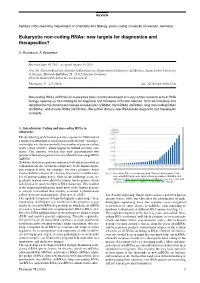
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Secretariat of the CBD Technical Series No. 82 Convention on Biological Diversity
Secretariat of the CBD Technical Series No. 82 Convention on Biological Diversity 82 SYNTHETIC BIOLOGY FOREWORD To be added by SCBD at a later stage. 1 BACKGROUND 2 In decision X/13, the Conference of the Parties invited Parties, other Governments and relevant 3 organizations to submit information on, inter alia, synthetic biology for consideration by the Subsidiary 4 Body on Scientific, Technical and Technological Advice (SBSTTA), in accordance with the procedures 5 outlined in decision IX/29, while applying the precautionary approach to the field release of synthetic 6 life, cell or genome into the environment. 7 Following the consideration of information on synthetic biology during the sixteenth meeting of the 8 SBSTTA, the Conference of the Parties, in decision XI/11, noting the need to consider the potential 9 positive and negative impacts of components, organisms and products resulting from synthetic biology 10 techniques on the conservation and sustainable use of biodiversity, requested the Executive Secretary 11 to invite the submission of additional relevant information on this matter in a compiled and synthesised 12 manner. The Secretariat was also requested to consider possible gaps and overlaps with the applicable 13 provisions of the Convention, its Protocols and other relevant agreements. A synthesis of this 14 information was thus prepared, peer-reviewed and subsequently considered by the eighteenth meeting 15 of the SBSTTA. The documents were then further revised on the basis of comments from the SBSTTA 16 and peer review process, and submitted for consideration by the twelfth meeting of the Conference of 17 the Parties to the Convention on Biological Diversity. -

3.1.4 Droplet-Based Microfluidics
Rodriguez Garcia, Marc (2016) Engineering the transition from non-living to living matter. PhD thesis. https://theses.gla.ac.uk/7605/ Copyright and moral rights for this work are retained by the author A copy can be downloaded for personal non-commercial research or study, without prior permission or charge This work cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given Enlighten: Theses https://theses.gla.ac.uk/ [email protected] Engineering the transition from non-living to living matter Marc Rodríguez Garcia A thesis submitted to the University of Glasgow for the degree of Doctor of Philosophy School of Chemistry College of Science and Engineering August 2016 A tota la meva família, per haver-me ajudat a arribar fins aquí. Però sobretot als meus pares, per estar tant a prop malgrat la distància. I en especial a la Nuria, per ser el motiu que em fa tirar endavant. “The good thing about science is that it's true whether or not you believe in it.” -Neil deGrasse Tyson Acknowledgements 1 Acknowledgements This project was carried out between September 2012 and June 2016 in the group of Prof Leroy Cronin in the School of Chemistry at the University of Glasgow. I have received much help and support from many colleagues and friends. -

Chapter 23 Nucleic Acids
7-9/99 Neuman Chapter 23 Chapter 23 Nucleic Acids from Organic Chemistry by Robert C. Neuman, Jr. Professor of Chemistry, emeritus University of California, Riverside [email protected] <http://web.chem.ucsb.edu/~neuman/orgchembyneuman/> Chapter Outline of the Book ************************************************************************************** I. Foundations 1. Organic Molecules and Chemical Bonding 2. Alkanes and Cycloalkanes 3. Haloalkanes, Alcohols, Ethers, and Amines 4. Stereochemistry 5. Organic Spectrometry II. Reactions, Mechanisms, Multiple Bonds 6. Organic Reactions *(Not yet Posted) 7. Reactions of Haloalkanes, Alcohols, and Amines. Nucleophilic Substitution 8. Alkenes and Alkynes 9. Formation of Alkenes and Alkynes. Elimination Reactions 10. Alkenes and Alkynes. Addition Reactions 11. Free Radical Addition and Substitution Reactions III. Conjugation, Electronic Effects, Carbonyl Groups 12. Conjugated and Aromatic Molecules 13. Carbonyl Compounds. Ketones, Aldehydes, and Carboxylic Acids 14. Substituent Effects 15. Carbonyl Compounds. Esters, Amides, and Related Molecules IV. Carbonyl and Pericyclic Reactions and Mechanisms 16. Carbonyl Compounds. Addition and Substitution Reactions 17. Oxidation and Reduction Reactions 18. Reactions of Enolate Ions and Enols 19. Cyclization and Pericyclic Reactions *(Not yet Posted) V. Bioorganic Compounds 20. Carbohydrates 21. Lipids 22. Peptides, Proteins, and α−Amino Acids 23. Nucleic Acids ************************************************************************************** -

Searching for Nucleic Acid Alternatives
MODIFIED OLIGONUCLEOTIDES 836 CHIMIA 2005, 59, No. 11 Chimia 59 (2005) 836–850 © Schweizerische Chemische Gesellschaft ISSN 0009–4293 Searching for Nucleic Acid Alternatives Albert Eschenmoser* Abstract: “Back of the envelope” methods have their place in experimental chemical research; they are effective mediators in the generation of research ideas, for instance, the design of molecular structures. Their qualitative character is part of their strength, rather than a drawback for the role they have to play. Qualitative conformational analysis of oligonucleotide and other oligomer systems on the level of idealized conformations is one such method; it has played a helpful role in our work on the chemical etiology of nucleic acid structure. This article, while giving a short overview of that work, shows how. Keywords: Conformational analysis of oligonucleotides · Nucleic acid analogs · Oligonucleodipeptides · p-RNA · TNA · Watson-Crick base-pairing Chemists understand by comparing, not ‘ab structural or transformational complexity, we know today as the molecular basis of initio’. To perceive and to create opportuni- serve the purpose of creating opportunities genetic function. The specific property to ties for drawing conclusions on the basis of to compare the behavior of complex sys- be compared in this work is a given nucleic comparisons is the organic chemist’s way tems with that of simpler ones. Enzymic re- acid alternative’s capacity for informational of interpreting and exploring the world at actions and enzyme models are examples. -

Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation
G C A T T A C G G C A T genes Review Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation Christian Diwo 1 and Nediljko Budisa 1,2,* 1 Institut für Chemie, Technische Universität Berlin Müller-Breslau-Straße 10, 10623 Berlin, Germany; [email protected] 2 Department of Chemistry, University of Manitoba, 144 Dysart Rd, 360 Parker Building, Winnipeg, MB R3T 2N2, Canada * Correspondence: [email protected] or [email protected]; Tel.: +49-30-314-28821 or +1-204-474-9178 Received: 27 November 2018; Accepted: 21 December 2018; Published: 28 December 2018 Abstract: The universal genetic code, which is the foundation of cellular organization for almost all organisms, has fostered the exchange of genetic information from very different paths of evolution. The result of this communication network of potentially beneficial traits can be observed as modern biodiversity. Today, the genetic modification techniques of synthetic biology allow for the design of specialized organisms and their employment as tools, creating an artificial biodiversity based on the same universal genetic code. As there is no natural barrier towards the proliferation of genetic information which confers an advantage for a certain species, the naturally evolved genetic pool could be irreversibly altered if modified genetic information is exchanged. We argue that an alien genetic code which is incompatible with nature is likely to assure the inhibition of all mechanisms of genetic information transfer in an open environment. The two conceivable routes to synthetic life are either de novo cellular design or the successive alienation of a complex biological organism through laboratory evolution. -

1 History for Canonical and Non-Canonical Structures of Nucleic Acids
1 1 History for Canonical and Non-canonical Structures of Nucleic Acids The main points of the learning: Understand canonical and non-canonical structures of nucleic acids and think of historical scientists in the research field of nucleic acids. 1.1 Introduction This book is to interpret the non-canonical structures and their stabilities of nucleic acids from the viewpoint of the chemistry and study their biological significances. There is more than 60 years’ history after the discovery of the double helix DNA structure by James Dewey Watson and Francis Harry Compton Crick in 1953, and chemical biology of nucleic acids is facing a new aspect today. Through this book, I expect that readers understand how the uncommon structure of nucleic acids became one of the common structures that fascinate us now. In this chapter, I introduce the history of nucleic acid structures and the perspective of research for non-canonical nucleic acid structures (see also Chapter 15). 1.2 History of Duplex The opening of the history of genetics was mainly done by three researchers. Charles Robert Darwin, who was a scientist of natural science, pioneered genetics. The proposition of genetic concept is indicated in his book On the Origin of Species published in 1859. He indicated the theory of biological evolution, which is the basic scientific hypothesis of natural diversity. In other words, he proposed biological evolution, which changed among individuals by adapting to the environment and be passed on to the next generation. However, that was still a primitive idea for the genetic concept. After that, Gregor Johann Mendel, who was a priest in Brno, Czech Republic, confirmed the mechanism of gene evolution by using “factor” inherited from parent to children using pea plant in 1865.