Enhancement of Deoxyribozyme Activity by Cationic Copolymers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme
University of Central Florida STARS HIM 1990-2015 2015 Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme Amanda J. Cox University of Central Florida Part of the Biochemistry Commons Find similar works at: https://stars.library.ucf.edu/honorstheses1990-2015 University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by STARS. It has been accepted for inclusion in HIM 1990-2015 by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Cox, Amanda J., "Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme" (2015). HIM 1990-2015. 1861. https://stars.library.ucf.edu/honorstheses1990-2015/1861 USING ANTENNA TILE-ASSISTED SUBSTRATE DELIVERY TO IMPROVE THE DETECTION LIMITS OF DEOXYRIBOZYME BIOSENSORS by AMANDA J. COX A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Chemistry, Biochemistry Track in the College of Sciences and in the Burnett Honors College at the University of Central Florida Orlando, Florida Fall Term, 2015 Thesis Chair: Dr. Dmitry Kolpashchikov, PhD ABSTRACT One common limitation of enzymatic reactions is the diffusion of a substrate to the enzyme active site and/or the release of the reaction products. These reactions are known as diffusion – controlled. Overcoming this limitation may enable faster catalytic rates, which in the case of catalytic biosensors can potentially lower limits of detection of specific analyte. Here we created an artificial system to enable deoxyribozyme (Dz) 10-23 based biosensor to overcome its diffusion limit. -
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
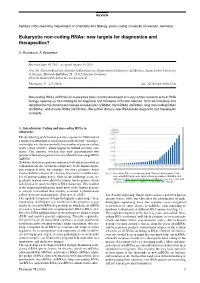
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Chapter 23 Nucleic Acids
7-9/99 Neuman Chapter 23 Chapter 23 Nucleic Acids from Organic Chemistry by Robert C. Neuman, Jr. Professor of Chemistry, emeritus University of California, Riverside [email protected] <http://web.chem.ucsb.edu/~neuman/orgchembyneuman/> Chapter Outline of the Book ************************************************************************************** I. Foundations 1. Organic Molecules and Chemical Bonding 2. Alkanes and Cycloalkanes 3. Haloalkanes, Alcohols, Ethers, and Amines 4. Stereochemistry 5. Organic Spectrometry II. Reactions, Mechanisms, Multiple Bonds 6. Organic Reactions *(Not yet Posted) 7. Reactions of Haloalkanes, Alcohols, and Amines. Nucleophilic Substitution 8. Alkenes and Alkynes 9. Formation of Alkenes and Alkynes. Elimination Reactions 10. Alkenes and Alkynes. Addition Reactions 11. Free Radical Addition and Substitution Reactions III. Conjugation, Electronic Effects, Carbonyl Groups 12. Conjugated and Aromatic Molecules 13. Carbonyl Compounds. Ketones, Aldehydes, and Carboxylic Acids 14. Substituent Effects 15. Carbonyl Compounds. Esters, Amides, and Related Molecules IV. Carbonyl and Pericyclic Reactions and Mechanisms 16. Carbonyl Compounds. Addition and Substitution Reactions 17. Oxidation and Reduction Reactions 18. Reactions of Enolate Ions and Enols 19. Cyclization and Pericyclic Reactions *(Not yet Posted) V. Bioorganic Compounds 20. Carbohydrates 21. Lipids 22. Peptides, Proteins, and α−Amino Acids 23. Nucleic Acids ************************************************************************************** -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Synthetic Biology Applying Engineering to Biology
Synthetic Biology Applying Engineering to Biology Report of a NEST High-Level Expert Group EUR 21796 PROJECT REPORT Interested in European research? RTD info is our quarterly magazine keeping you in touch with main developments (results, programmes, events, etc). It is available in English, French and German. A free sample copy or free subscription can be obtained from: European Commission Directorate-General for Research Information and Communication Unit B-1049 Brussels Fax : (32-2) 29-58220 E-mail: [email protected] Internet: http://europa.eu.int/comm/research/rtdinfo/index_en.html EUROPEAN COMMISSION Directorate-General for Research Directorate B — Structuring the European Research Area Unit B1 — Anticipation of Scientific and Technological Needs (NEST activity); Basic Research E-mail: [email protected] Contact: Christian Krassnig European Commission Office SDME 01/37 B-1049 Brussels Tel. (32-2) 29-86445 Fax (32-2) 29-93173 E-mail: [email protected] For further information on the NEST activity please refer to the following website: http://www.cordis.lu/nest/home.html EUROPEAN COMMISSION Synthetic Biology Applying Engineering to Biology Report of a NEST High-Level Expert Group NEST - New and Energing Science and Technology - is a research activity under the European Community’s 6th Framework Programme Directorate-General for Research Structuring the European Research Area 2005 Anticipating Scientific and Technological Needs; Basic Research EUR 21796 Europe Direct is a service to help you find answers to your questions about the European Union Freephone number: 00 800 6 7 8 9 10 11 LEGAL NOTICE: Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use which might be made of the following information. -

Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation
G C A T T A C G G C A T genes Review Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation Christian Diwo 1 and Nediljko Budisa 1,2,* 1 Institut für Chemie, Technische Universität Berlin Müller-Breslau-Straße 10, 10623 Berlin, Germany; [email protected] 2 Department of Chemistry, University of Manitoba, 144 Dysart Rd, 360 Parker Building, Winnipeg, MB R3T 2N2, Canada * Correspondence: [email protected] or [email protected]; Tel.: +49-30-314-28821 or +1-204-474-9178 Received: 27 November 2018; Accepted: 21 December 2018; Published: 28 December 2018 Abstract: The universal genetic code, which is the foundation of cellular organization for almost all organisms, has fostered the exchange of genetic information from very different paths of evolution. The result of this communication network of potentially beneficial traits can be observed as modern biodiversity. Today, the genetic modification techniques of synthetic biology allow for the design of specialized organisms and their employment as tools, creating an artificial biodiversity based on the same universal genetic code. As there is no natural barrier towards the proliferation of genetic information which confers an advantage for a certain species, the naturally evolved genetic pool could be irreversibly altered if modified genetic information is exchanged. We argue that an alien genetic code which is incompatible with nature is likely to assure the inhibition of all mechanisms of genetic information transfer in an open environment. The two conceivable routes to synthetic life are either de novo cellular design or the successive alienation of a complex biological organism through laboratory evolution. -

(12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub
US 20040O86860A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub. Date: May 6, 2004 (54) METHODS OF PRODUCING RNAS OF Publication Classification DEFINED LENGTH AND SEQUENCE (51) Int. Cl." .............................. C12Q 1/68; C12P 19/34 (76) Inventor: Muhammad Sohail, Marston (GB) (52) U.S. Cl. ............................................... 435/6; 435/91.2 Correspondence Address: MINTZ, LEVIN, COHN, FERRIS, GLOWSKY (57) ABSTRACT AND POPEO, PC. ONE FINANCIAL CENTER Methods of making RNA duplexes and single-stranded BOSTON, MA 02111 (US) RNAS of a desired length and Sequence based on cleavage of RNA molecules at a defined position, most preferably (21) Appl. No.: 10/264,748 with the use of deoxyribozymes. Novel deoxyribozymes capable of cleaving RNAS including a leader Sequence at a (22) Filed: Oct. 4, 2002 Site 3' to the leader Sequence are also described. Patent Application Publication May 6, 2004 Sheet 1 of 2 US 2004/0086860 A1 DNA Oligonucleotides T7 Promoter -TN-- OR 2N-2-N-to y Transcription Products GGGCGAAT-N-UU GGGCGAAT-N-UU w N Deoxyribozyme Cleavage - Q GGGCGAAT -------' Racction GGGCGAAT N-- UU N-UU ssRNA products N-UU Anneal ssRNA UU S-2N- UU siRNA product FIGURE 1: Flowchart summarising the procedure for siRNA synthesis. Patent Application Publication May 6, 2004 Sheet 2 of 2 US 2004/0086860 A1 Full-length transcript 3'-digestion product 5'-digestion product (5'GGGCGAATA) A: Production of single-stranded RNA templates by in vitro transcription and digestion With a deoxyribozyme V 2- 2 V 22inv 22 * 2 &3 S/AS - 88.8x, *...* or as IGFR -- is as 4. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Biosynthesis of Ribosylthymine in the Transfer RNA of Streptococcus
Proc. Nat. Acad. Sci. USA Vol. 72, No. 2, pp. 528-530, February 1975 Biosynthesis of Ribosylthymine in the Transfer RNA of Streptococcus faecalis: A Folate-Dependent Methylation Not Involving S-Adenosylmethionine (thymine/tRNA modification/formate/[methyl-'4Clmethionine/GlT'C-loop) ANN S. DELK AND JESSE C. RABINOWITZ Department of Biochemistry, University of California, Berkeley, Calif. 94720 Communicated by Bruce Ames, November 18, 1974 ABSTRACT Ribosylthymine is not present in tRNA of to produce thymidine 5'-monophosphate, which is sub- Streptococcus faecalis grown in the absence of folic acid, sequently used in DNA synthesis. Because of the folate although this methylated residue does occur in the tRNA of this organism when it is grown in the presence of folate. requirement for T formation in S. faecalis, it appeared possible We have found that, unlike other methylated residues in that methylation of RNA in this organism, particularly in RNA of S. faecalis and other organisms, the methyl moiety the biosynthesis of T, involves a folate derivative and not of ribosylthymine of the tRNA of folate-sufficient S. S-adenosylmethionine. faecalis is not derived from S-adenosylmethionine. Pre- liminary evidence suggests that a folate derivative serves MATERIALS AND METHODS as the methyl donor in this methylation. Unless stated otherwise, S. faecalis (ATCC 8043) and Esche- When grown in the presence of folic acid, Streptococcus richia coli (MRE 600) were grown for several generations in faecalis, like other prokaryotes, initiates protein synthesis -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group.