Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nucleoside Phosphate-Conjugates Come of Age: Catalytic Transformation, Polymerase Recognition and Antiviral Properties.# Elisabetta Groaz* and Piet Herdewijn
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Lirias Send Orders for Reprints to [email protected] Current Medicinal Chemistry, Year, Volume 1 Nucleoside Phosphate-Conjugates Come of Age: Catalytic Transformation, Polymerase Recognition and Antiviral Properties.# Elisabetta Groaz* and Piet Herdewijn Medicinal Chemistry, Rega Institute for Medical Research, KU Leuven, Minderbroedersstraat 10, 3000 Leuven, Belgium Abstract: Over the past few decades, different types of nucleoside phosphate-conjugates have been under extensive investigation due to their favorable molecular lability with interesting catalytic hydrolysis mechanisms, recognition as polymerase substrates, and especially for their development as antiviral/anticancer protide therapeutics. The antiviral conjugates such as nucleoside phosphoesters and phosphoramidates that were discovered and developed in the initial years have been well reviewed by the pioneers in the field. In the present review, we will discuss the basic chemical and biological principles behind consideration of some representative structural classes. We will also summarize the chemical and biological properties of some of the more recent analogues that were synthesized and evaluated in our laboratory and by others. This includes new principles for their application as direct substrates of polymerases, nucleobase-dependent catalytic and antiviral activity, and a plausible ‘prodrug of a prodrug’ strategy for tissue/organ-specific targeted drug delivery. Keywords: Prodrug – Delivery – Stability – Polymerase - Phosphoramidates INTRODUCTION profile of these molecules through the “tuning” of the nature of the existing ligands and the pursuit of new protecting The ability of unnatural nucleoside-based therapeutics to moieties relying on chemical rather than enzymatic induce inhibition of uncontrolled cell proliferation or viral dependant activation mechanisms. -

Pentose PO4 Pathway, Fructose, Galactose Metabolism.Pptx
Pentose PO4 pathway, Fructose, galactose metabolism The Entner Doudoroff pathway begins with hexokinase producing Glucose 6 PO4 , but produce only one ATP. This pathway prevalent in anaerobes such as Pseudomonas, they doe not have a Phosphofructokinase. The pentose phosphate pathway (also called the phosphogluconate pathway and the hexose monophosphate shunt) is a biochemical pathway parallel to glycolysis that generates NADPH and pentoses. While it does involve oxidation of glucose, its primary role is anabolic rather than catabolic. There are two distinct phases in the pathway. The first is the oxidative phase, in which NADPH is generated, and the second is the non-oxidative synthesis of 5-carbon sugars. For most organisms, the pentose phosphate pathway takes place in the cytosol. For each mole of glucose 6 PO4 metabolized to ribulose 5 PO4, 2 moles of NADPH are produced. 6-Phosphogluconate dh is not only an oxidation step but it’s also a decarboxylation reaction. The primary results of the pathway are: The generation of reducing equivalents, in the form of NADPH, used in reductive biosynthesis reactions within cells (e.g. fatty acid synthesis). Production of ribose-5-phosphate (R5P), used in the synthesis of nucleotides and nucleic acids. Production of erythrose-4-phosphate (E4P), used in the synthesis of aromatic amino acids. Transketolase and transaldolase reactions are similar in that they transfer between carbon chains, transketolases 2 carbon units or transaldolases 3 carbon units. Regulation; Glucose-6-phosphate dehydrogenase is the rate- controlling enzyme of this pathway. It is allosterically stimulated by NADP+. The ratio of NADPH:NADP+ is normally about 100:1 in liver cytosol. -

Depression of Thymidylate Synthetase Activity in Response to Cytosine Arabinoside1
[CANCER RESEARCH 32, 1160-1169, June 1972] Depression of Thymidylate Synthetase Activity in Response To Cytosine Arabinoside1 DeWayne Roberts and Ellen V. Loehr St. Jude Children's Research Hospital, Memphis, Tennessee 38101 ¡D.R., E. V. L.J and Department of Pharmacology, University of Tennessee Medical Units, Memphis, Tennessee 38101 [D. R.J SUMMARY Cytosine arabinoside induces remission of acute granulocytic leukemia (11). After the administration of Depression of thymidylate synthetase activity by cytosine cytosine arabinoside to patients, changes in the enzyme arabinoside in CCRF-CEM cells was correlated with a blocking activity pattern of leukemic leukocytes occur (32). After drug of precursor incorporation into RNA and with cell lysis. With administration, a decrease in thymidylate synthetase activity is increasing concentrations of cytosine arabinoside in the observed as early as 1 hr and persists for 24 hr in some culture media, a proportional decrease of uridine patients. A concentration-dependent decrease in thymidylate incorporation into RNA and decrease in thymidylate synthetase activity also follows the addition of cytosine synthetase activity occurred and was accompanied by lysis of arabinoside to CCRF-CEM cultures (33). the cells. These effects continued to develop with drug Methotrexate elevates thymidylate synthetase activity in concentrations in excess of that required to block thymidine leukocytes from patients with acute leukemia (32), in rat liver incorporation into DNA. The addition of deoxycytidine at and transplantable tumors (27), and in cells of LI210 or concentrations that failed to reverse inhibition by cytosine CCRF-CEM cultures (32, 33). The simultaneous addition of arabinoside of thymidine incorporation into DNA reversed the cytosine arabinoside and methotrexate to CCRF-CEM cultures drug effects on uridine incorporation, thymidylate synthetase modulated the effect of each drug on thymidylate synthetase activity, and cell lysis. -

REDUCTION of PURINE CONTENT in COMMONLY CONSUMED MEAT PRODUCTS THROUGH RINSING and COOKING by Anna Ellington (Under the Directio
REDUCTION OF PURINE CONTENT IN COMMONLY CONSUMED MEAT PRODUCTS THROUGH RINSING AND COOKING by Anna Ellington (Under the direction of Yen-Con Hung) Abstract The commonly consumed meat products ground beef, ground turkey, and bacon were analyzed for purine content before and after a rinsing treatment. The rinsing treatment involved rinsing the meat samples using a wrist shaker in 5:1 ratio water: sample for 2 or 5 minutes then draining or centrifuging to remove water. The total purine content of 25% fat ground beef significantly decreased (p<0.05) from 8.58 mg/g protein to a range of 5.17-7.26 mg/g protein after rinsing treatments. After rinsing and cooking an even greater decrease was seen ranging from 4.59-6.32 mg/g protein. The total purine content of 7% fat ground beef significantly decreased from 7.80 mg/g protein to a range of 5.07-5.59 mg/g protein after rinsing treatments. A greater reduction was seen after rinsing and cooking in the range of 4.38-5.52 mg/g protein. Ground turkey samples showed no significant changes after rinsing, but significant decreases were seen after rinsing and cooking. Bacon samples showed significant decreases from 6.06 mg/g protein to 4.72 and 4.49 after 2 and 5 minute rinsing and to 4.53 and 4.68 mg/g protein after 2 and 5 minute rinsing and cooking. Overall, this study showed that rinsing foods in water effectively reduces total purine content and subsequent cooking after rinsing results in an even greater reduction of total purine content. -
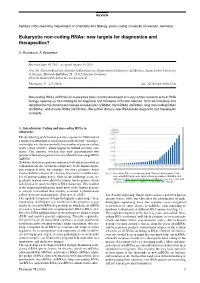
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Chapter 23 Nucleic Acids
7-9/99 Neuman Chapter 23 Chapter 23 Nucleic Acids from Organic Chemistry by Robert C. Neuman, Jr. Professor of Chemistry, emeritus University of California, Riverside [email protected] <http://web.chem.ucsb.edu/~neuman/orgchembyneuman/> Chapter Outline of the Book ************************************************************************************** I. Foundations 1. Organic Molecules and Chemical Bonding 2. Alkanes and Cycloalkanes 3. Haloalkanes, Alcohols, Ethers, and Amines 4. Stereochemistry 5. Organic Spectrometry II. Reactions, Mechanisms, Multiple Bonds 6. Organic Reactions *(Not yet Posted) 7. Reactions of Haloalkanes, Alcohols, and Amines. Nucleophilic Substitution 8. Alkenes and Alkynes 9. Formation of Alkenes and Alkynes. Elimination Reactions 10. Alkenes and Alkynes. Addition Reactions 11. Free Radical Addition and Substitution Reactions III. Conjugation, Electronic Effects, Carbonyl Groups 12. Conjugated and Aromatic Molecules 13. Carbonyl Compounds. Ketones, Aldehydes, and Carboxylic Acids 14. Substituent Effects 15. Carbonyl Compounds. Esters, Amides, and Related Molecules IV. Carbonyl and Pericyclic Reactions and Mechanisms 16. Carbonyl Compounds. Addition and Substitution Reactions 17. Oxidation and Reduction Reactions 18. Reactions of Enolate Ions and Enols 19. Cyclization and Pericyclic Reactions *(Not yet Posted) V. Bioorganic Compounds 20. Carbohydrates 21. Lipids 22. Peptides, Proteins, and α−Amino Acids 23. Nucleic Acids ************************************************************************************** -

2'-Deoxyguanosine Toxicity for B and Mature T Lymphoid Cell Lines Is Mediated by Guanine Ribonucleotide Accumulation
2'-deoxyguanosine toxicity for B and mature T lymphoid cell lines is mediated by guanine ribonucleotide accumulation. Y Sidi, B S Mitchell J Clin Invest. 1984;74(5):1640-1648. https://doi.org/10.1172/JCI111580. Research Article Inherited deficiency of the enzyme purine nucleoside phosphorylase (PNP) results in selective and severe T lymphocyte depletion which is mediated by its substrate, 2'-deoxyguanosine. This observation provides a rationale for the use of PNP inhibitors as selective T cell immunosuppressive agents. We have studied the relative effects of the PNP inhibitor 8- aminoguanosine on the metabolism and growth of lymphoid cell lines of T and B cell origin. We have found that 2'- deoxyguanosine toxicity for T lymphoblasts is markedly potentiated by 8-aminoguanosine and is mediated by the accumulation of deoxyguanosine triphosphate. In contrast, the growth of T4+ mature T cell lines and B lymphoblast cell lines is inhibited by somewhat higher concentrations of 2'-deoxyguanosine (ID50 20 and 18 microM, respectively) in the presence of 8-aminoguanosine without an increase in deoxyguanosine triphosphate levels. Cytotoxicity correlates instead with a three- to fivefold increase in guanosine triphosphate (GTP) levels after 24 h. Accumulation of GTP and growth inhibition also result from exposure to guanosine, but not to guanine at equimolar concentrations. B lymphoblasts which are deficient in the purine salvage enzyme hypoxanthine guanine phosphoribosyltransferase are completely resistant to 2'-deoxyguanosine or guanosine concentrations up to 800 microM and do not demonstrate an increase in GTP levels. Growth inhibition and GTP accumulation are prevented by hypoxanthine or adenine, but not by 2'-deoxycytidine. -

We Have Previously Reported' the Isolation of Guanosine Diphosphate
VOL. 48, 1962 BIOCHEMISTRY: HEATH AND ELBEIN 1209 9 Ramel, A., E. Stellwagen, and H. K. Schachman, Federation Proc., 20, 387 (1961). 10 Markus, G., A. L. Grossberg, and D. Pressman, Arch. Biochem. Biophys., 96, 63 (1962). "1 For preparation of anti-Xp antisera, see Nisonoff, A., and D. Pressman, J. Immunol., 80, 417 (1958) and idem., 83, 138 (1959). 12 For preparation of anti-Ap antisera, see Grossberg, A. L., and D. Pressman, J. Am. Chem. Soc., 82, 5478 (1960). 13 For preparation of anti-Rp antisera, see Pressman, D. and L. A. Sternberger, J. Immunol., 66, 609 (1951), and Grossberg, A. L., G. Radzimski, and D. Pressman, Biochemistry, 1, 391 (1962). 14 Smithies, O., Biochem. J., 71, 585 (1959). 15 Poulik, M. D., Biochim. et Biophysica Acta., 44, 390 (1960). 16 Edelman, G. M., and M. D. Poulik, J. Exp. Med., 113, 861 (1961). 17 Breinl, F., and F. Haurowitz, Z. Physiol. Chem., 192, 45 (1930). 18 Pauling, L., J. Am. Chem. Soc., 62, 2643 (1940). 19 Pressman, D., and 0. Roholt, these PROCEEDINGS, 47, 1606 (1961). THE ENZYMATIC SYNTHESIS OF GUANOSINE DIPHOSPHATE COLITOSE BY A MUTANT STRAIN OF ESCHERICHIA COLI* BY EDWARD C. HEATHt AND ALAN D. ELBEINT RACKHAM ARTHRITIS RESEARCH UNIT AND DEPARTMENT OF BACTERIOLOGY, THE UNIVERSITY OF MICHIGAN Communicated by J. L. Oncley, May 10, 1962 We have previously reported' the isolation of guanosine diphosphate colitose (GDP-colitose* GDP-3,6-dideoxy-L-galactose) from Escherichia coli 0111-B4; only 2.5 umoles of this sugar nucleotide were isolated from 1 kilogram of cells. Studies on the biosynthesis of colitose with extracts of this organism indicated that GDP-mannose was a precursor;2 however, the enzymatically formed colitose was isolated from a high-molecular weight substance and attempts to isolate the sus- pected intermediate, GDP-colitose, were unsuccessful. -

Lecture 7 - the Calvin Cycle and the Pentose Phosphate Pathway
Lecture 7 - The Calvin Cycle and the Pentose Phosphate Pathway Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction The Calvin cycle Text The dark reactions of photosynthesis in green plants Reduces carbon from CO2 to hexose (C6H12O6) Requires ATP for free energy and NADPH as a reducing agent. 2 2 Introduction NADH versus Text NADPH 3 3 Introduction The Pentose Phosphate Pathway Used in all organisms Glucose is oxidized and decarboxylated to produce reduced NADPH Used for the synthesis and degradation of pentoses Shares reactions with the Calvin cycle 4 4 1. The Calvin Cycle Source of carbon is CO2 Text Takes place in the stroma of the chloroplasts Comprises three stages Fixation of CO2 by ribulose 1,5-bisphosphate to form two 3-phosphoglycerate molecules Reduction of 3-phosphoglycerate to produce hexose sugars Regeneration of ribulose 1,5-bisphosphate 5 5 1. Calvin Cycle Three stages 6 6 1.1 Stage I: Fixation Incorporation of CO2 into 3-phosphoglycerate 7 7 1.1 Stage I: Fixation Rubisco: Ribulose 1,5- bisphosphate carboxylase/ oxygenase 8 8 1.1 Stage I: Fixation Active site contains a divalent metal ion 9 9 1.2 Rubisco Oxygenase Activity Rubisco also catalyzes a wasteful oxygenase reaction: 10 10 1.3 State II: Formation of Hexoses Reactions similar to those of gluconeogenesis But they take place in the chloroplasts And use NADPH instead of NADH 11 11 1.3 State III: Regeneration of Ribulose 1,5-Bisphosphosphate Involves a sequence of transketolase and aldolase reactions. 12 12 1.3 State III: -

An Introduction to Recurrent Nucleotide Interactions in RNA Blake A
Overview An introduction to recurrent nucleotide interactions in RNA Blake A. Sweeney,1 Poorna Roy2 and Neocles B. Leontis2∗ RNA secondary structure diagrams familiar to molecular biologists summarize at a glance the folding of RNA chains to form Watson–Crick paired double helices. However, they can be misleading: First of all, they imply that the nucleotides in loops and linker segments, which can amount to 35% to 50% of a structured RNA, do not significantly interact with other nucleotides. Secondly, they give the impression that RNA molecules are loosely organized in three-dimensional (3D) space. In fact, structured RNAs are compactly folded as a result of numerous long-range, sequence-specific interactions, many of which involve loop or linker nucleotides. Here, we provide an introduction for students and researchers of RNA on the types, prevalence, and sequence variations of inter-nucleotide interactions that structure and stabilize RNA 3D motifs and architectures, using Escherichia coli (E. coli) 16S ribosomal RNA as a concrete example. The picture that emerges is that almost all nucleotides in structured RNA molecules, including those in nominally single-stranded loop or linker regions, form specific interactions that stabilize functional structures or mediate interactions with other molecules. The small number of noninteracting, ‘looped-out’ nucleotides make it possible for the RNA chain to form sharp turns. Base-pairing is the most specific interaction in RNA as it involves edge-to-edge hydrogen bonding (H-bonding) of the bases. Non-Watson–Crick base pairs are a significant fraction (30% or more) of base pairs in structured RNAs. © 2014 John Wiley & Sons, Ltd. -

Inhibition by Cyclic Guanosine 3':5'-Monophosphate of the Soluble DNA Polymerase Activity, and of Partially Purified DNA Polymer
Inhibition by Cyclic Guanosine 3':5'-Monophosphate of the Soluble DNA Polymerase Activity, and of Partially Purified DNA Polymerase A (DNA Polymerase I) from the Yeast Saccharomyces cere visiae Hans Eckstein Institut für Physiologische Chemie der Universität, Martinistr. 52-UKE, D-2000 Hamburg 20 Z. Naturforsch. 36 c, 813-819 (1981); received April 16/July 2, 1981 Dedicated to Professor Dr. Joachim Kühnauon the Occasion of His 80th Birthday cGMP, DNA Polymerase Activity, DNA Polymerase A, DNA Polymerase I, Baker’s Yeast DNA polymerase activity from extracts of growing yeast cells is inhibited by cGMP. Experiments with partially purified yeast DNA polymerases show, that cGMP inhibits DNA polymerase A (DNA polymerase I from Chang), which is the main component of the soluble DNA polymerase activity in yeast extracts, by competing for the enzyme with the primer- template DNA. Since the enzyme is not only inhibited by 3',5'-cGMP, but also by 3',5'-cAMP, the 3': 5'-phosphodiester seems to be crucial for the competition between cGMP and primer. This would be inconsistent with the concept of a 3'-OH primer binding site in the enzyme. The existence of such a site in the yeast DNA polymerase A is indicated from studies with various purine nucleoside monophosphates. When various DNA polymerases are compared, inhibition by cGMP seems to be restricted to those enzymes, which are involved in DNA replication. DNA polymerases with an associated nuclease activity are not inhibited, DNA polymerase B from yeast is even activated by cGMP. Though some relations between the cGMP effect and the presumed function of the enzymes in the living cell are apparent, the biological meaning of the observations in general remains open. -

Reversibility Ofthe Pyrophosphoryl Transfer from ATP to GTP By
Proc. Nat. Acad. Sci. USA Vol. 71, No. 9, pp. 3470-3473, September 1974 Reversibility of the Pyrophosphoryl Transfer from ATP to GTP by Escherichia coli Stringent Factor (guanosine 5'-diphosphate-3'-diphosphate/guanosine 5'-triphosphate-3'-diphosphate/relaxed control/ribosomes) JOSE SY The Rockefeller University, New York, N.Y. 10021 Communicated by Fritz Lipmann, July 1, 1974 ABSTRACT The stringent factor-catalyzed, ribosome- was incubated for 60 min at 300 in a 1W0-1 mixture contain- dependent synthesis of guanosine polyplosphates is ing: 50 mM Tris OAc, pH 8.1; 4 mM dithiothreitol; 20 mM found to be reversible. The reverse reaction specifically requires 5'-AMP as the pyrophosphoryl acceptor, and Mg(OAc)2; 5 mM ATP; 10 ,ug of poly(A, U, G); 27 pgof tRNA; guanosine 5'-triphosphate-3'-diphospbate is preferentially 90',g of ethanol-washed ribosomes; and 2 p&g of fraction II utilized as the pyrophosphoryl donor. The primary prod- stringent factor. By minimizing GTPase activity with the ucts of the reaction are GTP and ANP. The reverse reaction use of high Mg++ concentration and ethanol-washed ribo- is strongly inhibited by the antibiotics thiostrepton and a ob- tetracycline, and b' A P and,1-y-pnethylene-adenosixie- somes with a minimal time of incubation, product is triphosphate, but not by ADP, GTP, and GIDP. The reverse tained that is more than 95% pppGpp. After incubation, 1 Al reaction occurs under conditions for nonribosomal syn- of 88% HCOOH was added and the resulting precipitate dis- thesis. The ove'rill reaction for stringent factor-catalyzed carded.