Introduction to Bioinformatics Introduction to Molecular Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Discovery of DNA Structure and Function: Watson and Crick By: Leslie A
01/08/2018 Discovery of DNA Double Helix: Watson and Crick | Learn Science at Scitable NUCLEIC ACID STRUCTURE AND FUNCTION | Lead Editor: Bob Moss Discovery of DNA Structure and Function: Watson and Crick By: Leslie A. Pray, Ph.D. © 2008 Nature Education Citation: Pray, L. (2008) Discovery of DNA structure and function: Watson and Crick. Nature Education 1(1):100 The landmark ideas of Watson and Crick relied heavily on the work of other scientists. What did the duo actually discover? Aa Aa Aa Many people believe that American biologist James Watson and English physicist Francis Crick discovered DNA in the 1950s. In reality, this is not the case. Rather, DNA was first identified in the late 1860s by Swiss chemist Friedrich Miescher. Then, in the decades following Miescher's discovery, other scientists--notably, Phoebus Levene and Erwin Chargaff--carried out a series of research efforts that revealed additional details about the DNA molecule, including its primary chemical components and the ways in which they joined with one another. Without the scientific foundation provided by these pioneers, Watson and Crick may never have reached their groundbreaking conclusion of 1953: that the DNA molecule exists in the form of a three-dimensional double helix. The First Piece of the Puzzle: Miescher Discovers DNA Although few people realize it, 1869 was a landmark year in genetic research, because it was the year in which Swiss physiological chemist Friedrich Miescher first identified what he called "nuclein" inside the nuclei of human white blood cells. (The term "nuclein" was later changed to "nucleic acid" and eventually to "deoxyribonucleic acid," or "DNA.") Miescher's plan was to isolate and characterize not the nuclein (which nobody at that time realized existed) but instead the protein components of leukocytes (white blood cells). -

A Correlation of Cytological and Genetical Crossing-Over in Zea Mays. PNAS 17:492–497
A CORRELATION OF CYTOLOGICAL AND GENETICAL CROSSING-OVER IN ZEA MAYS HARRIET B. CREIGHTON BARBARA MCCLINTOCK Botany Department Cornell University Ithaca, New York Creighton, H., and McClintock, B. 1931 A correlation of cytological and genetical crossing-over in Zea mays. PNAS 17:492–497. E S P Electronic Scholarly Publishing http://www.esp.org Electronic Scholarly Publishing Project Foundations Series –– Classical Genetics Series Editor: Robert J. Robbins The ESP Foundations of Classical Genetics project has received support from the ELSI component of the United States Department of Energy Human Genome Project. ESP also welcomes help from volunteers and collaborators, who recommend works for publication, provide access to original materials, and assist with technical and production work. If you are interested in volunteering, or are otherwise interested in the project, contact the series editor: [email protected]. Bibliographical Note This ESP edition, first electronically published in 2003 and subsequently revised in 2018, is a newly typeset, unabridged version, based on the 1931 edition published by The National Academy of Sciences. Unless explicitly noted, all footnotes and endnotes are as they appeared in the original work. Some of the graphics have been redone for this electronic version. Production Credits Scanning of originals: ESP staff OCRing of originals: ESP staff Typesetting: ESP staff Proofreading/Copyediting: ESP staff Graphics work: ESP staff Copyfitting/Final production: ESP staff © 2003, 2018 Electronic Scholarly Publishing Project http://www.esp.org This electronic edition is made freely available for educational or scholarly purposes, provided that this copyright notice is included. The manuscript may not be reprinted or redistributed for commercial purposes without permission. -
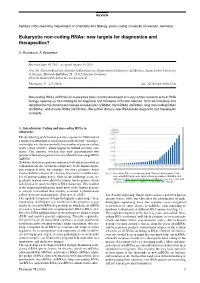
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Introduction and Historical Perspective
Chapter 1 Introduction and Historical Perspective “ Nothing in biology makes sense except in the light of evolution. ” modified by the developmental history of the organism, Theodosius Dobzhansky its physiology – from cellular to systems levels – and by the social and physical environment. Finally, behaviors are shaped through evolutionary forces of natural selection OVERVIEW that optimize survival and reproduction ( Figure 1.1 ). Truly, the study of behavior provides us with a window through Behavioral genetics aims to understand the genetic which we can view much of biology. mechanisms that enable the nervous system to direct Understanding behaviors requires a multidisciplinary appropriate interactions between organisms and their perspective, with regulation of gene expression at its core. social and physical environments. Early scientific The emerging field of behavioral genetics is still taking explorations of animal behavior defined the fields shape and its boundaries are still being defined. Behavioral of experimental psychology and classical ethology. genetics has evolved through the merger of experimental Behavioral genetics has emerged as an interdisciplin- psychology and classical ethology with evolutionary biol- ary science at the interface of experimental psychology, ogy and genetics, and also incorporates aspects of neuro- classical ethology, genetics, and neuroscience. This science ( Figure 1.2 ). To gain a perspective on the current chapter provides a brief overview of the emergence of definition of this field, it is helpful -

DNA: the Timeline and Evidence of Discovery
1/19/2017 DNA: The Timeline and Evidence of Discovery Interactive Click and Learn (Ann Brokaw Rocky River High School) Introduction For almost a century, many scientists paved the way to the ultimate discovery of DNA and its double helix structure. Without the work of these pioneering scientists, Watson and Crick may never have made their ground-breaking double helix model, published in 1953. The knowledge of how genetic material is stored and copied in this molecule gave rise to a new way of looking at and manipulating biological processes, called molecular biology. The breakthrough changed the face of biology and our lives forever. Watch The Double Helix short film (approximately 15 minutes) – hyperlinked here. 1 1/19/2017 1865 The Garden Pea 1865 The Garden Pea In 1865, Gregor Mendel established the foundation of genetics by unraveling the basic principles of heredity, though his work would not be recognized as “revolutionary” until after his death. By studying the common garden pea plant, Mendel demonstrated the inheritance of “discrete units” and introduced the idea that the inheritance of these units from generation to generation follows particular patterns. These patterns are now referred to as the “Laws of Mendelian Inheritance.” 2 1/19/2017 1869 The Isolation of “Nuclein” 1869 Isolated Nuclein Friedrich Miescher, a Swiss researcher, noticed an unknown precipitate in his work with white blood cells. Upon isolating the material, he noted that it resisted protein-digesting enzymes. Why is it important that the material was not digested by the enzymes? Further work led him to the discovery that the substance contained carbon, hydrogen, nitrogen and large amounts of phosphorus with no sulfur. -

Jewels in the Crown
Jewels in the crown CSHL’s 8 Nobel laureates Eight scientists who have worked at Cold Max Delbrück and Salvador Luria Spring Harbor Laboratory over its first 125 years have earned the ultimate Beginning in 1941, two scientists, both refugees of European honor, the Nobel Prize for Physiology fascism, began spending their summers doing research at Cold or Medicine. Some have been full- Spring Harbor. In this idyllic setting, the pair—who had full-time time faculty members; others came appointments elsewhere—explored the deep mystery of genetics to the Lab to do summer research by exploiting the simplicity of tiny viruses called bacteriophages, or a postdoctoral fellowship. Two, or phages, which infect bacteria. Max Delbrück and Salvador who performed experiments at Luria, original protagonists in what came to be called the Phage the Lab as part of the historic Group, were at the center of a movement whose members made Phage Group, later served as seminal discoveries that launched the revolutionary field of mo- Directors. lecular genetics. Their distinctive math- and physics-oriented ap- Peter Tarr proach to biology, partly a reflection of Delbrück’s physics train- ing, was propagated far and wide via the famous Phage Course that Delbrück first taught in 1945. The famous Luria-Delbrück experiment of 1943 showed that genetic mutations occur ran- domly in bacteria, not necessarily in response to selection. The pair also showed that resistance was a heritable trait in the tiny organisms. Delbrück and Luria, along with Alfred Hershey, were awarded a Nobel Prize in 1969 “for their discoveries concerning the replication mechanism and the genetic structure of viruses.” Barbara McClintock Alfred Hershey Today we know that “jumping genes”—transposable elements (TEs)—are littered everywhere, like so much Alfred Hershey first came to Cold Spring Harbor to participate in Phage Group wreckage, in the chromosomes of every organism. -

Genes, Genomes and Genetic Analysis
© Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION UNIT 1 © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FORDefining SALE OR DISTRIBUTION and WorkingNOT FOR SALE OR DISTRIBUTION with Genes © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION Chapter 1 Genes, Genomes, and Genetic Analysis Chapter 2 DNA Structure and Genetic Variation © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Molekuul/Science Photo Library/Getty Images. © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC. NOT FOR SALE OR DISTRIBUTION 9781284136609_CH01_Hartl.indd 1 08/11/17 8:50 am © Jones & Bartlett Learning, LLC -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

MCDB 5220 Methods and Logics April 21 2015 Marcelo Bassalo
Cracking the Genetic Code MCDB 5220 Methods and Logics April 21 2015 Marcelo Bassalo The DNA Saga… so far Important contributions for cracking the genetic code: • The “transforming principle” (1928) Frederick Griffith The DNA Saga… so far Important contributions for cracking the genetic code: • The “transforming principle” (1928) • The nature of the transforming principle: DNA (1944 - 1952) Oswald Avery Alfred Hershey Martha Chase The DNA Saga… so far Important contributions for cracking the genetic code: • The “transforming principle” (1928) • The nature of the transforming principle: DNA (1944 - 1952) • X-ray diffraction and the structure of proteins (1951) Linus Carl Pauling The DNA Saga… so far Important contributions for cracking the genetic code: • The “transforming principle” (1928) • The nature of the transforming principle: DNA (1944 - 1952) • X-ray diffraction and the structure of proteins (1951) • The structure of DNA (1953) James Watson and Francis Crick The DNA Saga… so far Important contributions for cracking the genetic code: • The “transforming principle” (1928) • The nature of the transforming principle: DNA (1944 - 1952) • X-ray diffraction and the structure of proteins (1951) • The structure of DNA (1953) How is DNA (4 nucleotides) the genetic material while proteins (20 amino acids) are the building blocks? ? DNA Protein ? The Coding Craze ? DNA Protein What was already known? • DNA resides inside the nucleus - DNA is not the carrier • Protein synthesis occur in the cytoplasm through ribosomes {• Only RNA is associated with ribosomes (no DNA) - rRNA is not the carrier { • Ribosomal RNA (rRNA) was a homogeneous population The “messenger RNA” hypothesis François Jacob Jacques Monod The Coding Craze ? DNA RNA Protein RNA Tie Club Table from Wikipedia The Coding Craze Who won the race Marshall Nirenberg J. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Chromosomal Theory and Genetic Linkage
362 Chapter 13 | Modern Understandings of Inheritance 13.1 | Chromosomal Theory and Genetic Linkage By the end of this section, you will be able to do the following: • Discuss Sutton’s Chromosomal Theory of Inheritance • Describe genetic linkage • Explain the process of homologous recombination, or crossing over • Describe chromosome creation • Calculate the distances between three genes on a chromosome using a three-point test cross Long before scientists visualized chromosomes under a microscope, the father of modern genetics, Gregor Mendel, began studying heredity in 1843. With improved microscopic techniques during the late 1800s, cell biologists could stain and visualize subcellular structures with dyes and observe their actions during cell division and meiosis. With each mitotic division, chromosomes replicated, condensed from an amorphous (no constant shape) nuclear mass into distinct X-shaped bodies (pairs of identical sister chromatids), and migrated to separate cellular poles. Chromosomal Theory of Inheritance The speculation that chromosomes might be the key to understanding heredity led several scientists to examine Mendel’s publications and reevaluate his model in terms of chromosome behavior during mitosis and meiosis. In 1902, Theodor Boveri observed that proper sea urchin embryonic development does not occur unless chromosomes are present. That same year, Walter Sutton observed chromosome separation into daughter cells during meiosis (Figure 13.2). Together, these observations led to the Chromosomal Theory of Inheritance, which identified chromosomes as the genetic material responsible for Mendelian inheritance. Figure 13.2 (a) Walter Sutton and (b) Theodor Boveri developed the Chromosomal Theory of Inheritance, which states that chromosomes carry the unit of heredity (genes). -

In 1953 in England James Watson and Francis Crick Discovered the Structure of DNA in the Now-Famous Scientific Narrative Known As the “Race Towards the Double Helix”
THE NARRATIVES OF SCIENCE: LITERARY THEORY AND DISCOVERY IN MOLECULAR BIOLOGY PRIYA VENKATESAN In 1953 in England James Watson and Francis Crick discovered the structure of DNA in the now-famous scientific narrative known as the “race towards the double helix”. Meanwhile in France, Roland Barthes published his first book, Writing Degree Zero, on literary theory, which became the intellectual precursor for the new human sciences that were developing based on Saussurean linguistics. The discovery by Watson and Crick of the double helix marked a definitive turning point in the development of the life sciences, paving the way for the articulation of the genetic code and the emergence of molecular biology. The publication by Barthes was no less significant, since it served as an exemplar for elucidating how literary narratives are structured and for formulating how textual material is constructed. As Françoise Dosse notes, Writing Degree Zero “received unanimous acclaim and quickly became a symptom of new literary demands, a break with tradition”.1 Both the work of Roland Barthes and Watson and Crick served as paradigms in their respective fields. Semiotics, the field of textual analysis as developed by Barthes in Writing Degree Zero, offered a new direction in the structuring of narrative whereby each distinct unit in a story formed a “code” or “isotopy” that categorizes the formal elements of the story. The historical concurrence of the discovery of the double helix and the publication of Writing Degree Zero may be mere coincidence, but this essay is an exploration of the intellectual influence that both events may have had on each other, since both the discovery of the double helix and Barthes’ publication gave expression to the new forms of knowledge 1 Françoise Dosse, History of Structuralism: The Rising Sign, 1945-1966, trans.