Design and Switch of Catalytic Activity with the Dnazyme–Rnazyme Combination
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
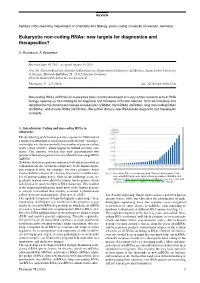
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

How Might a Pre-Biotic Ribozyme Catalyze RNA Assembly in an RNA World?
Science Highlight – April 2007 How Might a Pre-biotic Ribozyme Catalyze RNA Assembly in an RNA World? Which came first, nucleic acids or proteins? This question is molecular biology's version of the "chicken-or-the-egg" riddle. Genes made of nucleic acids (DNA or RNA) contain the instructions for making proteins, but enzymes made of proteins are needed to replicate genes. For those who try to understand how life origi- nated, this once seemed an intractable paradox. The discovery 25 years ago that RNA can be enzymatic permits us to speculate that pre-biotic self-replicating molecules may have been RNAs (1,2). This is known as the "RNA World" hypothesis, and with the discovery of RNA catalysis, it is now possible to imagine a prebiotic The L1 Ligase ribozyme at the moment 'RNA World' (or even one populated by early life forms) of bond creation. in which self-replicating ribozymes (RNA-based enzymes that possess the catalytic ability to copy themselves) accomplished both tasks, thus avoiding the potential “chicken-or-the-egg” conundrum (3). But there's a catch. In order to copy RNA, fragments or monomers that have 5'-triphosphates must be ligated together. This is true for modern polymerases, and is also the most likely mechanism by which a ribozyme self-replicase in an RNA World might function. Yet no one has found a modern natural ribozyme that catalyze this The RNA nucleotide triphosphate ligation reaction required reaction (pictured right). for RNA polymerization and self-replication. RNA in vitro evolution and selection has however enabled several research groups to discover RNA sequences that can in fact cata- lyze the required chemical reaction (shown above) for 5'-triphos- phate RNA fragment ligation, and one group has even produced a primitive but functional RNA-based RNA polymerase ribozyme (4). -

A Tetracycline-Dependent Ribozyme Switch Allows Conditional Induction of Gene Expression in Caenorhabditis Elegans
ARTICLE https://doi.org/10.1038/s41467-019-08412-w OPEN A tetracycline-dependent ribozyme switch allows conditional induction of gene expression in Caenorhabditis elegans Lena A. Wurmthaler1,2, Monika Sack1,2, Karina Gense2,3, Jörg S. Hartig1,2 & Martin Gamerdinger2,3 The nematode Caenorhabditis elegans represents an important research model. Convenient methods for conditional induction of gene expression in this organism are not available. Here 1234567890():,; we describe tetracycline-dependent ribozymes as versatile RNA-based genetic switches in C. elegans. Ribozyme insertion into the 3’-UTR converts any gene of interest into a tetracycline- inducible gene allowing temporal and, by using tissue-selective promoters, spatial control of expression in all developmental stages of the worm. Using the ribozyme switches we established inducible C. elegans polyglutamine Huntington’s disease models exhibiting ligand- controlled polyQ-huntingtin expression, inclusion body formation, and toxicity. Our approach circumvents the complicated expression of regulatory proteins. Moreover, only little coding space is necessary and natural promoters can be utilized. With these advantages tetracycline-dependent ribozymes significantly expand the genetic toolbox for C. elegans. 1 Department of Chemistry, University of Konstanz, 78457 Konstanz, Germany. 2 Konstanz Research School Chemical Biology (KoRS-CB), University of Konstanz, 78457 Konstanz, Germany. 3 Department of Biology, University of Konstanz, 78457 Konstanz, Germany. Correspondence and requests for materials should be addressed to J.S.H. (email: [email protected]) or to M.G. (email: [email protected]) NATURE COMMUNICATIONS | (2019) 10:491 | https://doi.org/10.1038/s41467-019-08412-w | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-019-08412-w nducible regulatory systems are very powerful research tools to single A-to-G point mutation in the catalytic core of the HHR9 Iinvestigate the cellular function of individual genes. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme
CHAPTER SEVEN Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme Sara M. O’Rourke, William G. Scott1 The Center for the Molecular Biology of RNA, University of California at Santa Cruz, Santa Cruz, CA, United States 1Corresponding author: e-mail address: [email protected] Contents 1. Background and Structural Overview 178 2. Fast Minimal Hammerhead Ribozymes 181 3. Acid-Base Catalysis and the Hammerhead Ribozyme 182 4. Is the Hammerhead Ligation Reaction the Reverse of the Cleavage Reaction? 191 5. Do Cooperative Interactions in the Hammerhead Ribozyme Facilitate General Base Catalysis in the Cleavage Reaction? 194 6. Summary and Concluding Remarks 195 6.1 The Structure of the Hammerhead Ribozyme May Be Much Simpler Than We Have Thought 196 6.2 The Mechanism of the Hammerhead Ribozyme May Be Much More Complicated Than We Have Thought 196 6.3 Concluding Remarks 200 References 201 Abstract Natural or full-length hammerhead ribozymes are up to 1000-fold more active than their minimal counterparts that lack a complex tertiary interaction that pre-organizes and stabilizes the ribozyme active site, positioning RNA functional groups to facilitate acid-base catalysis. The recent discovery that a single tertiary contact (an AU Hoogsteen pair) between Stems I and II confers essentially all of the enhanced activity greatly simplifies our understanding of the structural requirements for hammerhead ribozyme activity. In contrast, the simplest mechanistic interpretations are challenged with the presentation of more complex alternatives. These alternatives are elucidated and criti- cally analyzed in the context of several of the active hammerhead ribozyme structures now available. # Progress in Molecular Biology and Translational Science, Volume 159 2018 Elsevier Inc. -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group. -

Ribozymes Targeted to the Mitochondria Using the 5S Ribosomal Rna
RIBOZYMES TARGETED TO THE MITOCHONDRIA USING THE 5S RIBOSOMAL RNA By JENNIFER ANN BONGORNO A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Jennifer Bongorno To my grandmother, Hazel Traster Miller, whose interest in genealogy sparked my interest in genetics, and without whose mitochondria I would not be here ACKNOWLEDGMENTS I would like to thank all the members of the Lewin lab; especially my mentor, Al Lewin. Al was always there for me with suggestions and keeping me motivated. He and the other members of the lab were like my second family; I would not have had an enjoyable experience without them. Diana Levinson and Elizabeth Bongorno worked with me on the fourth and third mouse transfections respectively. Joe Hartwich and Al Lewin tested some of the ribozymes in vitro and cloned some of the constructs I used. James Thomas also helped with cloning and was an invaluable lab manager. Verline Justilien worked on a related project and was a productive person with whom to bounce ideas back and forth. Lourdes Andino taught me how to use the new phosphorimager for my SYBR Green-stained gels. Alan White was there through it all, like the older brother I never had. Mary Ann Checkley was with me even longer than Alan, since we both came to Florida from Ohio Wesleyan, although she did manage to graduate before me. Jia Liu and Frederic Manfredsson were there when I needed a beer. -

Mirna Detection Using a Rolling Circle Amplification and RNA
biosensors Article MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy Chenxin Fang, Ping Ouyang, Yuxing Yang, Yang Qing, Jialun Han, Wenyan Shang, Yubing Chen and Jie Du * State Key Laboratory of Marine Resource Utilization in South China Sea, College of Materials Science and Engineering, Hainan University, Haikou 570228, China; [email protected] (C.F.); [email protected] (P.O.); [email protected] (Y.Y.); [email protected] (Y.Q.); [email protected] (J.H.); [email protected] (W.S.); [email protected] (Y.C.) * Correspondence: [email protected] Abstract: A microRNA (miRNA) detection platform composed of a rolling circle amplification (RCA) system and an allosteric deoxyribozyme system is proposed, which can detect miRNA-21 rapidly and efficiently. Padlock probe hybridization with the target miRNA is achieved through complementary base pairing and the padlock probe forms a closed circular template under the action of ligase; this circular template results in RCA. In the presence of DNA polymerase, RCA proceeds and a long chain with numerous repeating units is formed. In the presence of single-stranded DNA (H1 and H2), multi-component nucleic acid enzymes (MNAzymes) are formed that have the ability to cleave substrates. Finally, substrates containing fluorescent and quenching groups and magnesium ions are added to the system to activate the MNAzyme and the substrate cleavage reaction, thus achieving fluorescence intensity amplification. The RCA–MNAzyme system has dual signal amplification and presents a sensing platform that demonstrates broad prospects in the analysis and detection of Citation: Fang, C.; Ouyang, P.; Yang, nucleic acids. -

Neo-Cloo2 B2 O 1 B 1 (3) REVERSE TRANSCRIPTION RNA PROCESSING
|||||||||||||| O US005436141A United States Patent (19) 11 Patent Number: 5,436,141 Miyata et al. (45) Date of Patent: Jul. 25, 1995 54 METHOD FOR SYNTHESIZING STABLE 56) References Cited SINGLE-STRANDED CONAN EUKARYOTES BY MEANS OF A U.S. PATENT DOCUMENTS BACTERAL RETRON AND PRODUCTS 5,079,151 1/1992 Lampson et al. ................ 435/91.51 75 Inventors: Shohei Miyata, Misato, Japan; FOREIGN PATENT DOCUMENTS Atsushi Ohshima, Highland Park, 0132309 1/1985 European Pat. Off. ... C12N 15/00 N.J.; Sumiko Inouye; Masayori Inouye, both of Bridgewater, N.J. OTHER PUBLICATIONS Y o Dhundale et al., J. Bact, vol. 170, 1988, pp. 5620-5624. 73) Assignee: University of Medicine and Dentistry Lampson et al., Science, vol. 243, 1989, pp. 1033-1038. of New Jersey, Newark, N.J. Sambrook et al., Molecular Cloning: A Laboratory Man ual, Cold Spring Harbor Laboratory Press, 1989, pp. 21 Appl. No.: 753,110 16.15-16.16. 22 Filed: Aug. 30, 1991 Primary Examiner-Richard A. Schwartz Assistant Examiner-James Ketter Related U.S. Application Data Attorney, Agent, or Firm-Weiser & Associates (63) Continuation-in-part of Ser. No. 315,427, Feb. 24, 1989, 57 ABSTRACT Pat. No. 5,979,151, and a continuation in part of Ser. A method for producing in vivo stable single-stranded No. 315,316, Feb. 24, 1989, Pat. No. 5,320,958, and a DNAs in eucaryotic cells. The DNAs are multicopy Eup ii.g 1. single-stranded DNA (msDNA) structures constituted 517,946, May 2, 1990, and a continuation-in-part of Ser. by a RNA and a DNA portion. -

One Sequence, Two Ribozymes: Could Access by Neutral Drift Every Sequence on Both Networks
R EPORTS minus the background levels observed in the HSP in X-100 for 15 min at 4¡C with intermittent mixing, 62. M. R. Peterson, C. G. Burd, S. D. Emr, Curr. Biol. 9, 159 the control (Sar1-GDPÐcontaining) incubation that and elutes were pooled. Proteins were precipitated by (1999). prevents COPII vesicle formation. In the microsome MeOH/CH3Cl and separated by SDSÐpolyacrylamide 63. M. G. Waters, D. O. Clary, J. E. Rothman, J. Cell Biol. control, the level of p115-SNARE associations was gel electrophoresis (PAGE) followed by immunoblot- 118, 1015 (1992). less than 0.1%. ting using p115 mAb 13F12. 64. D. M. Walter, K. S. Paul, M. G. Waters, J. Biol. Chem. 46. C. M. Carr, E. Grote, M. Munson, F. M. Hughson, P. J. 51. V. Rybin et al., Nature 383, 266 (1996). 273, 29565 (1998). Novick, J. Cell Biol. 146, 333 (1999). 52. K. G. Hardwick and H. R. Pelham, J. Cell Biol. 119, 513 65. N. Hui et al., Mol. Biol. Cell 8, 1777 (1997). 47. C. Ungermann, B. J. Nichols, H. R. Pelham, W. Wick- (1992). 66. T. E. Kreis, EMBO J. 5, 931 (1986). ner, J. Cell Biol. 140, 61 (1998). 53. A. P. Newman, M. E. Groesch, S. Ferro-Novick, EMBO 67. H. Plutner, H. W. Davidson, J. Saraste, W. E. Balch, 48. E. Grote and P. J. Novick, Mol. Biol. Cell 10, 4149 J. 11, 3609 (1992). J. Cell Biol. 119, 1097 (1992). (1999). 54. A. Spang and R. Schekman, J. Cell Biol. 143, 589 (1998). 68. D. S. -

De Novo Nucleic Acids: a Review of Synthetic Alternatives to DNA and RNA That Could Act As † Bio-Information Storage Molecules
life Review De Novo Nucleic Acids: A Review of Synthetic Alternatives to DNA and RNA That Could Act as y Bio-Information Storage Molecules Kevin G Devine 1 and Sohan Jheeta 2,* 1 School of Human Sciences, London Metropolitan University, 166-220 Holloway Rd, London N7 8BD, UK; [email protected] 2 Network of Researchers on the Chemical Evolution of Life (NoR CEL), Leeds LS7 3RB, UK * Correspondence: [email protected] This paper is dedicated to Professor Colin B Reese, Daniell Professor of Chemistry, Kings College London, y on the occasion of his 90th Birthday. Received: 17 November 2020; Accepted: 9 December 2020; Published: 11 December 2020 Abstract: Modern terran life uses several essential biopolymers like nucleic acids, proteins and polysaccharides. The nucleic acids, DNA and RNA are arguably life’s most important, acting as the stores and translators of genetic information contained in their base sequences, which ultimately manifest themselves in the amino acid sequences of proteins. But just what is it about their structures; an aromatic heterocyclic base appended to a (five-atom ring) sugar-phosphate backbone that enables them to carry out these functions with such high fidelity? In the past three decades, leading chemists have created in their laboratories synthetic analogues of nucleic acids which differ from their natural counterparts in three key areas as follows: (a) replacement of the phosphate moiety with an uncharged analogue, (b) replacement of the pentose sugars ribose and deoxyribose with alternative acyclic, pentose and hexose derivatives and, finally, (c) replacement of the two heterocyclic base pairs adenine/thymine and guanine/cytosine with non-standard analogues that obey the Watson–Crick pairing rules.