The CPEB3 Ribozyme Modulates Hippocampal-Dependent Memory
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Germline-Specific Isoform of Eif4e (IFE-1) Is Required for Efficient Translation of Stored Mrnas and Maturation of Both Oocytes and Sperm
Research Article 1529 A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm Melissa A. Henderson1, Elizabeth Cronland1, Steve Dunkelbarger2, Vince Contreras1, Susan Strome2 and Brett D. Keiper1,* 1Department of Biochemistry and Molecular Biology, Brody School of Medicine at East Carolina University, Greenville, NC 27834, USA 2Department of Molecular Cell and Developmental Biology, University of California, Santa Cruz, CA 95064, USA *Author for correspondence (e-mail: [email protected]) Accepted 26 January 2009 Journal of Cell Science 122, 1529-1539 Published by The Company of Biologists 2009 doi:10.1242/jcs.046771 Summary Fertility and embryonic viability are measures of efficient germ CED-4/Apaf-1, and accumulated as multinucleate cells unable cell growth and development. During oogenesis and to mature to spermatids. A modest defect in oocyte development spermatogenesis, new proteins are required for both mitotic was also observed. Oocytes progressed normally through mitosis expansion and differentiation. Qualitative and quantitative and meiosis, but subsequent production of competent oocytes changes in protein synthesis occur by translational control of became limiting, even in the presence of wild-type sperm. mRNAs, mediated in part by eIF4E, which binds the mRNAs Combined gametogenesis defects decreased worm fertility by 5Ј cap. IFE-1 is one of five eIF4E isoforms identified in 80% at 20°C; ife-1 worms were completely sterile at 25°C. C. elegans. IFE-1 is expressed primarily in the germ line and Thus, IFE-1 plays independent roles in late oogenesis and associates with P granules, large mRNPs that store mRNAs. -

Vaccination with Prion Peptide-Displaying Polyomavirus-Like Particles Prolongs Incubation Time in Scrapie-Infected Mice
viruses Article Vaccination with Prion Peptide-Displaying Polyomavirus-Like Particles Prolongs Incubation Time in Scrapie-Infected Mice Martin Eiden 1,* , Alma Gedvilaite 2 , Fabienne Leidel 1,3, Rainer G. Ulrich 1 and Martin H. Groschup 1 1 Institute of Novel and Emerging Infectious Diseases, Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Südufer 10, 17493 Greifswald-Insel Riems, Germany; [email protected] (F.L.); rainer.ulrich@fli.de (R.G.U.); martin.groschup@fli.de (M.H.G.) 2 Life Sciences Center, Institute of Biotechnology, Vilnius University, Sauletekio˙ al. 7, LT-10257 Vilnius, Lithuania; [email protected] 3 Task Force Animal Diseases, Darmstadt Regional Administrative Council, Luisenplatz 2, 64283 Darmstadt, Germany * Correspondence: martin.eiden@fli.de Abstract: Prion diseases like scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle or Creutzfeldt–Jakob disease (CJD) in humans are fatal neurodegenerative diseases characterized by the conformational conversion of the normal, mainly α-helical cellular prion protein (PrPC) into the abnormal β-sheet rich infectious isoform PrPSc. Various therapeutic or prophylactic approaches have been conducted, but no approved therapeutic treatment is available so far. Immunisation against prions is hampered by the self-tolerance to PrPC in mammalian species. One strategy to avoid this tolerance is presenting PrP variants in virus-like particles (VLPs). Therefore, we vaccinated C57/BL6 mice with nine prion peptide variants presented by hamster polyomavirus capsid protein VP1/VP2-derived VLPs. Mice were subsequently challenged intraperitoneally with the murine RML Citation: Eiden, M.; Gedvilaite, A.; prion strain. Importantly, one group exhibited significantly increased mean survival time of 240 days Leidel, F.; Ulrich, R.G.; Groschup, M.H. -

How Might a Pre-Biotic Ribozyme Catalyze RNA Assembly in an RNA World?
Science Highlight – April 2007 How Might a Pre-biotic Ribozyme Catalyze RNA Assembly in an RNA World? Which came first, nucleic acids or proteins? This question is molecular biology's version of the "chicken-or-the-egg" riddle. Genes made of nucleic acids (DNA or RNA) contain the instructions for making proteins, but enzymes made of proteins are needed to replicate genes. For those who try to understand how life origi- nated, this once seemed an intractable paradox. The discovery 25 years ago that RNA can be enzymatic permits us to speculate that pre-biotic self-replicating molecules may have been RNAs (1,2). This is known as the "RNA World" hypothesis, and with the discovery of RNA catalysis, it is now possible to imagine a prebiotic The L1 Ligase ribozyme at the moment 'RNA World' (or even one populated by early life forms) of bond creation. in which self-replicating ribozymes (RNA-based enzymes that possess the catalytic ability to copy themselves) accomplished both tasks, thus avoiding the potential “chicken-or-the-egg” conundrum (3). But there's a catch. In order to copy RNA, fragments or monomers that have 5'-triphosphates must be ligated together. This is true for modern polymerases, and is also the most likely mechanism by which a ribozyme self-replicase in an RNA World might function. Yet no one has found a modern natural ribozyme that catalyze this The RNA nucleotide triphosphate ligation reaction required reaction (pictured right). for RNA polymerization and self-replication. RNA in vitro evolution and selection has however enabled several research groups to discover RNA sequences that can in fact cata- lyze the required chemical reaction (shown above) for 5'-triphos- phate RNA fragment ligation, and one group has even produced a primitive but functional RNA-based RNA polymerase ribozyme (4). -

Zinc and Copper Ions Differentially Regulate Prion-Like Phase
viruses Article Zinc and Copper Ions Differentially Regulate Prion-Like Phase Separation Dynamics of Pan-Virus Nucleocapsid Biomolecular Condensates Anne Monette 1,* and Andrew J. Mouland 1,2,* 1 Lady Davis Institute at the Jewish General Hospital, Montréal, QC H3T 1E2, Canada 2 Department of Medicine, McGill University, Montréal, QC H4A 3J1, Canada * Correspondence: [email protected] (A.M.); [email protected] (A.J.M.) Received: 2 September 2020; Accepted: 12 October 2020; Published: 18 October 2020 Abstract: Liquid-liquid phase separation (LLPS) is a rapidly growing research focus due to numerous demonstrations that many cellular proteins phase-separate to form biomolecular condensates (BMCs) that nucleate membraneless organelles (MLOs). A growing repertoire of mechanisms supporting BMC formation, composition, dynamics, and functions are becoming elucidated. BMCs are now appreciated as required for several steps of gene regulation, while their deregulation promotes pathological aggregates, such as stress granules (SGs) and insoluble irreversible plaques that are hallmarks of neurodegenerative diseases. Treatment of BMC-related diseases will greatly benefit from identification of therapeutics preventing pathological aggregates while sparing BMCs required for cellular functions. Numerous viruses that block SG assembly also utilize or engineer BMCs for their replication. While BMC formation first depends on prion-like disordered protein domains (PrLDs), metal ion-controlled RNA-binding domains (RBDs) also orchestrate their formation. Virus replication and viral genomic RNA (vRNA) packaging dynamics involving nucleocapsid (NC) proteins and their orthologs rely on Zinc (Zn) availability, while virus morphology and infectivity are negatively influenced by excess Copper (Cu). While virus infections modify physiological metal homeostasis towards an increased copper to zinc ratio (Cu/Zn), how and why they do this remains elusive. -

A Tetracycline-Dependent Ribozyme Switch Allows Conditional Induction of Gene Expression in Caenorhabditis Elegans
ARTICLE https://doi.org/10.1038/s41467-019-08412-w OPEN A tetracycline-dependent ribozyme switch allows conditional induction of gene expression in Caenorhabditis elegans Lena A. Wurmthaler1,2, Monika Sack1,2, Karina Gense2,3, Jörg S. Hartig1,2 & Martin Gamerdinger2,3 The nematode Caenorhabditis elegans represents an important research model. Convenient methods for conditional induction of gene expression in this organism are not available. Here 1234567890():,; we describe tetracycline-dependent ribozymes as versatile RNA-based genetic switches in C. elegans. Ribozyme insertion into the 3’-UTR converts any gene of interest into a tetracycline- inducible gene allowing temporal and, by using tissue-selective promoters, spatial control of expression in all developmental stages of the worm. Using the ribozyme switches we established inducible C. elegans polyglutamine Huntington’s disease models exhibiting ligand- controlled polyQ-huntingtin expression, inclusion body formation, and toxicity. Our approach circumvents the complicated expression of regulatory proteins. Moreover, only little coding space is necessary and natural promoters can be utilized. With these advantages tetracycline-dependent ribozymes significantly expand the genetic toolbox for C. elegans. 1 Department of Chemistry, University of Konstanz, 78457 Konstanz, Germany. 2 Konstanz Research School Chemical Biology (KoRS-CB), University of Konstanz, 78457 Konstanz, Germany. 3 Department of Biology, University of Konstanz, 78457 Konstanz, Germany. Correspondence and requests for materials should be addressed to J.S.H. (email: [email protected]) or to M.G. (email: [email protected]) NATURE COMMUNICATIONS | (2019) 10:491 | https://doi.org/10.1038/s41467-019-08412-w | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-019-08412-w nducible regulatory systems are very powerful research tools to single A-to-G point mutation in the catalytic core of the HHR9 Iinvestigate the cellular function of individual genes. -

Prion Diseases in Knock-In Mice Carrying Single Prp Codon Substitutions Associated with Human Diseases
Profoundly different prion diseases in knock-in mice carrying single PrP codon substitutions associated with human diseases Walker S. Jacksona,b,c,1, Andrew W. Borkowskia,b,c, Nicki E. Watsona, Oliver D. Kingd, Henryk Faase, Alan Jasanoffe,f, and Susan Lindquista,b,c,2 aWhitehead Institute for Biomedical Research, Cambridge, MA 02142; bHoward Hughes Medical Institute, cDepartment of Biology, and fDepartments of Biological Engineering, Brain and Cognitive Sciences, and Nuclear Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139; dDepartment of Cell and Developmental Biology, University of Massachusetts Medical School, Worcester, MA 01655; and eFrances Bitter Magnet Laboratory, Cambridge, MA 02139 Contributed by Susan Lindquist, July 9, 2013 (sent for review March 7, 2013) In man, mutations in different regions of the prion protein (PrP) linked to it, provides an important general model for such are associated with infectious neurodegenerative diseases that investigations. have remarkably different clinical signs and neuropathological There are several types of human prion diseases, each begin- lesions. To explore the roots of this phenomenon, we created ning with pathologic processes in a different brain region and fi – a knock-in mouse model carrying the mutation associated with leading to distinct functional de cits: cognition [Creutzfeldt – – one of these diseases [Creutzfeldt–Jakob disease (CJD)] that was Jakob disease (CJD)], movement control (Gerstmann Sträussler Scheinker syndrome), or sleep and autonomic functions [fatal exactly analogous to a previous knock-in model of a different fl prion disease [fatal familial insomnia (FFI)]. Together with the familial insomnia (FFI)] (7). Prion diseases also af ict animals WT parent, this created an allelic series of three lines, each express- and include bovine spongiform encephalopathy (BSE) of cattle, scrapie of sheep and goats, and chronic wasting disease (CWD) ing the same protein with a single amino acid difference, and with of deer and elk (1). -

Mitochondrial Dysfunction in Preclinical Genetic Prion Disease: a Target for 7 Preventive Treatment?
1HXURELRORJ\RI'LVHDVH ² Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Mitochondrial dysfunction in preclinical genetic prion disease: A target for 7 preventive treatment? Guy Kellera,c,1, Orli Binyamina,c,1, Kati Frida,c, Ann Saadab,c, Ruth Gabizona,c,⁎ a Department of Neurology, The Agnes Ginges Center for Human Neurogenetics, Israel b Department of Genetics and Metabolic Diseases, The Monique and Jacques Roboh Department of Genetic Research, Hadassah-Hebrew University Medical Center, Israel c Medical School, The Hebrew University, Jerusalem, Israel ABSTRACT Mitochondrial malfunction is a common feature in advanced stages of neurodegenerative conditions, as is the case for the accumulation of aberrantly folded proteins, such as PrP in prion diseases. In this work, we investigated mitochondrial activity and expression of related factors vis a vis PrP accumulation at the subclinical stages of TgMHu2ME199K mice, modeling for genetic prion diseases. While these mice remain healthy until 5–6 months of age, they succumb to fatal disease at 12–14 months. We found that mitochondrial respiratory chain enzymatic activates and ATP/ROS production, were abnormally elevated in asymptomatic mice, concomitant with initial accumulation of disease related PrP. In parallel, the expression of Cytochrome c oxidase (COX) subunit IV isoform 1(Cox IV-1) was reduced and replaced by the activity of Cox IV isoform 2, which operates in oxidative neuronal conditions. At all stages of disease, Cox IV-1 was absent from cells accu- mulating disease related PrP, suggesting that PrP aggregates may directly compromise normal mitochondrial function. Administration of Nano-PSO, a brain targeted antioxidant, to TgMHu2ME199K mice, reversed functional and biochemical mitochondrial functions to normal conditions regardless of the presence of misfolded PrP. -

An Evolutionary Basis for Scrapie Disease : Identification of a Fish
72 Update TRENDS in Genetics Vol.19 No.2 February 2003 affect regulation of the human genome in a more global 11 Frith, M.C. et al. (2002) Statistical significance of clusters of motifs manner by creating S/MARs that form chromatin loops, represented by position specific scoring matrices in nucleotide and by shaping the sequence evolution of LCRs. sequences. Nucleic Acids Res. 30, 3214–3224 12 Wingender, E. et al. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 29, 281–283 Acknowledgements 13 Purucker, M. et al. (1990) Structure and function of the enhancer 30 to Galina V. Glazko was supported by research grants from NIH (GM-20293) the human A gamma globin gene. Nucleic Acids Res. 18, 7407–7415 and NASA (NCC2-1057) awarded to Masatoshi Nei. We thank Nathan 14 Li, Q. et al. (1999) Locus control regions: coming of age at a decade plus. J. Bowen and Wolfgang J. Miller for discussions on the relationship Trends Genet. 15, 403–408 between TEs and S/MARs. 15 Hardison, R. et al. (1997) Locus control regions of mammalian beta- globin gene clusters: combining phylogenetic analyses and exper- References imental results to gain functional insights. Gene 205, 73–94 1 The Human Genome Sequencing Consortium, (2001) Initial sequen- 16 Strausberg, R. et al. (1999) The mammalian gene collection. Science cing and analysis of the human genome. Nature 409, 860–921 286, 455–457 2 Kidwell, M.G. and Lisch, D.R. (2001) Perspective: transposable 17 Bode, J. et al. (1996) Scaffold/matrix-attached regions: topological elements, parasitic DNA, and genome evolution. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme
CHAPTER SEVEN Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme Sara M. O’Rourke, William G. Scott1 The Center for the Molecular Biology of RNA, University of California at Santa Cruz, Santa Cruz, CA, United States 1Corresponding author: e-mail address: [email protected] Contents 1. Background and Structural Overview 178 2. Fast Minimal Hammerhead Ribozymes 181 3. Acid-Base Catalysis and the Hammerhead Ribozyme 182 4. Is the Hammerhead Ligation Reaction the Reverse of the Cleavage Reaction? 191 5. Do Cooperative Interactions in the Hammerhead Ribozyme Facilitate General Base Catalysis in the Cleavage Reaction? 194 6. Summary and Concluding Remarks 195 6.1 The Structure of the Hammerhead Ribozyme May Be Much Simpler Than We Have Thought 196 6.2 The Mechanism of the Hammerhead Ribozyme May Be Much More Complicated Than We Have Thought 196 6.3 Concluding Remarks 200 References 201 Abstract Natural or full-length hammerhead ribozymes are up to 1000-fold more active than their minimal counterparts that lack a complex tertiary interaction that pre-organizes and stabilizes the ribozyme active site, positioning RNA functional groups to facilitate acid-base catalysis. The recent discovery that a single tertiary contact (an AU Hoogsteen pair) between Stems I and II confers essentially all of the enhanced activity greatly simplifies our understanding of the structural requirements for hammerhead ribozyme activity. In contrast, the simplest mechanistic interpretations are challenged with the presentation of more complex alternatives. These alternatives are elucidated and criti- cally analyzed in the context of several of the active hammerhead ribozyme structures now available. # Progress in Molecular Biology and Translational Science, Volume 159 2018 Elsevier Inc. -
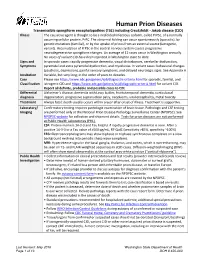
Prion Disease Reporting and Investigation Guideline
Human Prion Diseases Transmissible spongiform encephalopathies (TSE) including Creutzfeldt - Jakob disease (CJD) Illness The causative agent is thought to be a misfolded infectious isoform, called PrPSc, of a normally occurring cellular protein, PrPC. The abnormal folding can occur spontaneously (sporadic), by genetic mutations (familial), or by the uptake of prions from an external source (iatrogenic, variant). Accumulation of PrPSc in the central nervous system causes progressive neurodegenerative spongiform changes. An average of 12 cases occur in Washington annually. No cases of variant CJD have been reported in Washington state to date. Signs and In sporadic cases: rapidly progressive dementia, visual disturbances, cerebellar dysfunction, Symptoms pyramidal and extra pyramidal dysfunction, and myoclonus. In variant cases: behavioral changes (psychosis, depression), painful sensory symptoms, and delayed neurologic signs. See Appendix A Incubation Variable, but very long; in the order of years to decades. Case Please see https://www.cdc.gov/prions/cjd/diagnostic-criteria.html for sporadic, familial, and Classification iatrogenic CJD and https://www.cdc.gov/prions/vcjd/diagnostic-criteria.html for variant CJD. Report all definite, probable and possible cases to CDE Differential Alzheimer’s disease, dementia with Lewy bodies, frontotemporal dementia, corticobasal diagnosis degeneration, progressive supranuclear palsy, neoplasms, viral encephalitis, metal toxicity Treatment Always fatal; death usually occurs within a year after onset of illness. Treatment is supportive. Laboratory/ Confirmatory testing requires pathologic examination of brain tissue. Pathologic and CSF testing Imaging are performed only at the National Prion Disease Pathology Surveillance Center (NPDPSC). See NPDPSC website for collection and shipment details. Tests for prion diseases are not performed at Public Health Laboratories (PHL). -

Ribozymes Targeted to the Mitochondria Using the 5S Ribosomal Rna
RIBOZYMES TARGETED TO THE MITOCHONDRIA USING THE 5S RIBOSOMAL RNA By JENNIFER ANN BONGORNO A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Jennifer Bongorno To my grandmother, Hazel Traster Miller, whose interest in genealogy sparked my interest in genetics, and without whose mitochondria I would not be here ACKNOWLEDGMENTS I would like to thank all the members of the Lewin lab; especially my mentor, Al Lewin. Al was always there for me with suggestions and keeping me motivated. He and the other members of the lab were like my second family; I would not have had an enjoyable experience without them. Diana Levinson and Elizabeth Bongorno worked with me on the fourth and third mouse transfections respectively. Joe Hartwich and Al Lewin tested some of the ribozymes in vitro and cloned some of the constructs I used. James Thomas also helped with cloning and was an invaluable lab manager. Verline Justilien worked on a related project and was a productive person with whom to bounce ideas back and forth. Lourdes Andino taught me how to use the new phosphorimager for my SYBR Green-stained gels. Alan White was there through it all, like the older brother I never had. Mary Ann Checkley was with me even longer than Alan, since we both came to Florida from Ohio Wesleyan, although she did manage to graduate before me. Jia Liu and Frederic Manfredsson were there when I needed a beer.