Conditional Deoxyribozyme−Nanoparticle Conjugates For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme
University of Central Florida STARS HIM 1990-2015 2015 Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme Amanda J. Cox University of Central Florida Part of the Biochemistry Commons Find similar works at: https://stars.library.ucf.edu/honorstheses1990-2015 University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by STARS. It has been accepted for inclusion in HIM 1990-2015 by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Cox, Amanda J., "Using Antenna Tile-Assisted Substrate Delivery to Improve Detection Limits of Deoxyribozyme" (2015). HIM 1990-2015. 1861. https://stars.library.ucf.edu/honorstheses1990-2015/1861 USING ANTENNA TILE-ASSISTED SUBSTRATE DELIVERY TO IMPROVE THE DETECTION LIMITS OF DEOXYRIBOZYME BIOSENSORS by AMANDA J. COX A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Chemistry, Biochemistry Track in the College of Sciences and in the Burnett Honors College at the University of Central Florida Orlando, Florida Fall Term, 2015 Thesis Chair: Dr. Dmitry Kolpashchikov, PhD ABSTRACT One common limitation of enzymatic reactions is the diffusion of a substrate to the enzyme active site and/or the release of the reaction products. These reactions are known as diffusion – controlled. Overcoming this limitation may enable faster catalytic rates, which in the case of catalytic biosensors can potentially lower limits of detection of specific analyte. Here we created an artificial system to enable deoxyribozyme (Dz) 10-23 based biosensor to overcome its diffusion limit. -

Engineering Nucleic Acid Chemistry for Precise and Controllable CRISPR
Science Bulletin 64 (2019) 1841–1849 Contents lists available at ScienceDirect Science Bulletin journal homepage: www.elsevier.com/locate/scib Review Engineering nucleic acid chemistry for precise and controllable CRISPR/Cas9 genome editing ⇑ Weiqi Cai a,b, Ming Wang a,b, a Beijing National Laboratory for Molecular Sciences, Key Laboratory of Analytical Chemistry for Living Biosystems, CAS Research/Education Center for Excellence in Molecule Science, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China b University of Chinese Academy of Sciences, Beijing 100049, China article info abstract Article history: The clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (CRISPR/ Received 21 June 2019 Cas9) genome editing technology is revolutionizing our approach and capability to precisely manipulate Received in revised form 16 July 2019 the genetic flow of mammalians. The facile programmability of Cas9 protein and guide RNA (gRNA) Accepted 23 July 2019 sequence has recently expanded biomedical application of CRISPR/Cas9 technology from editing mam- Available online 30 July 2019 malian genome to various genetic manipulations. The therapeutic and clinical translation potential of CRISPR/Cas9 genome editing, however, are challenged by its off-target effect and low genome editing effi- Keywords: ciency. In this regard, developing new Cas9 variants and conditional control of Cas9/gRNA activity are of CRISPR/Cas9 genome editing great potential for improving genome editing accuracy and on-target efficiency. In this review, we sum- RNA engineering Nucleic acid chemistry marize chemical strategies that have been developed recently to engineer the nucleic acid chemistry of Gene therapy gRNA to enhance CRISPR/Cas9 genome editing efficacy, specificity and controllability. -
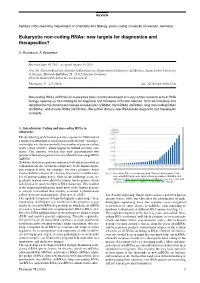
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Synthetic Biology Applying Engineering to Biology
Synthetic Biology Applying Engineering to Biology Report of a NEST High-Level Expert Group EUR 21796 PROJECT REPORT Interested in European research? RTD info is our quarterly magazine keeping you in touch with main developments (results, programmes, events, etc). It is available in English, French and German. A free sample copy or free subscription can be obtained from: European Commission Directorate-General for Research Information and Communication Unit B-1049 Brussels Fax : (32-2) 29-58220 E-mail: [email protected] Internet: http://europa.eu.int/comm/research/rtdinfo/index_en.html EUROPEAN COMMISSION Directorate-General for Research Directorate B — Structuring the European Research Area Unit B1 — Anticipation of Scientific and Technological Needs (NEST activity); Basic Research E-mail: [email protected] Contact: Christian Krassnig European Commission Office SDME 01/37 B-1049 Brussels Tel. (32-2) 29-86445 Fax (32-2) 29-93173 E-mail: [email protected] For further information on the NEST activity please refer to the following website: http://www.cordis.lu/nest/home.html EUROPEAN COMMISSION Synthetic Biology Applying Engineering to Biology Report of a NEST High-Level Expert Group NEST - New and Energing Science and Technology - is a research activity under the European Community’s 6th Framework Programme Directorate-General for Research Structuring the European Research Area 2005 Anticipating Scientific and Technological Needs; Basic Research EUR 21796 Europe Direct is a service to help you find answers to your questions about the European Union Freephone number: 00 800 6 7 8 9 10 11 LEGAL NOTICE: Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use which might be made of the following information. -

(12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub
US 20040O86860A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub. Date: May 6, 2004 (54) METHODS OF PRODUCING RNAS OF Publication Classification DEFINED LENGTH AND SEQUENCE (51) Int. Cl." .............................. C12Q 1/68; C12P 19/34 (76) Inventor: Muhammad Sohail, Marston (GB) (52) U.S. Cl. ............................................... 435/6; 435/91.2 Correspondence Address: MINTZ, LEVIN, COHN, FERRIS, GLOWSKY (57) ABSTRACT AND POPEO, PC. ONE FINANCIAL CENTER Methods of making RNA duplexes and single-stranded BOSTON, MA 02111 (US) RNAS of a desired length and Sequence based on cleavage of RNA molecules at a defined position, most preferably (21) Appl. No.: 10/264,748 with the use of deoxyribozymes. Novel deoxyribozymes capable of cleaving RNAS including a leader Sequence at a (22) Filed: Oct. 4, 2002 Site 3' to the leader Sequence are also described. Patent Application Publication May 6, 2004 Sheet 1 of 2 US 2004/0086860 A1 DNA Oligonucleotides T7 Promoter -TN-- OR 2N-2-N-to y Transcription Products GGGCGAAT-N-UU GGGCGAAT-N-UU w N Deoxyribozyme Cleavage - Q GGGCGAAT -------' Racction GGGCGAAT N-- UU N-UU ssRNA products N-UU Anneal ssRNA UU S-2N- UU siRNA product FIGURE 1: Flowchart summarising the procedure for siRNA synthesis. Patent Application Publication May 6, 2004 Sheet 2 of 2 US 2004/0086860 A1 Full-length transcript 3'-digestion product 5'-digestion product (5'GGGCGAATA) A: Production of single-stranded RNA templates by in vitro transcription and digestion With a deoxyribozyme V 2- 2 V 22inv 22 * 2 &3 S/AS - 88.8x, *...* or as IGFR -- is as 4. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Tissue-Specific Delivery of CRISPR Therapeutics
International Journal of Molecular Sciences Review Tissue-Specific Delivery of CRISPR Therapeutics: Strategies and Mechanisms of Non-Viral Vectors Karim Shalaby 1 , Mustapha Aouida 1,* and Omar El-Agnaf 1,2,* 1 Division of Biological and Biomedical Sciences (BBS), College of Health & Life Sciences (CHLS), Hamad Bin Khalifa University (HBKU), Doha 34110, Qatar; [email protected] 2 Neurological Disorders Research Center, Qatar Biomedical Research Institute (QBRI), Hamad Bin Khalifa University (HBKU), Doha 34110, Qatar * Correspondence: [email protected] (M.A.); [email protected] (O.E.-A.) Received: 12 September 2020; Accepted: 27 September 2020; Published: 5 October 2020 Abstract: The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) genome editing system has been the focus of intense research in the last decade due to its superior ability to desirably target and edit DNA sequences. The applicability of the CRISPR-Cas system to in vivo genome editing has acquired substantial credit for a future in vivo gene-based therapeutic. Challenges such as targeting the wrong tissue, undesirable genetic mutations, or immunogenic responses, need to be tackled before CRISPR-Cas systems can be translated for clinical use. Hence, there is an evident gap in the field for a strategy to enhance the specificity of delivery of CRISPR-Cas gene editing systems for in vivo applications. Current approaches using viral vectors do not address these main challenges and, therefore, strategies to develop non-viral delivery systems are being explored. Peptide-based systems represent an attractive approach to developing gene-based therapeutics due to their specificity of targeting, scale-up potential, lack of an immunogenic response and resistance to proteolysis. -

Ribozyme-Mediated, Multiplex CRISPR Gene Editing and Crispri in Plasmodium Yoelii
bioRxiv preprint doi: https://doi.org/10.1101/481416; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Ribozyme-Mediated, Multiplex CRISPR Gene Editing and CRISPRi in Plasmodium yoelii Michael P. Walker1 and Scott E. Lindner1 * 1Department of Biochemistry and Molecular Biology, the Huck Center for Malaria Research, Pennsylvania State University, University Park, PA. *Correspondence: Scott E. Lindner, [email protected] Running Title: CRISPR-RGR in Plasmodium yoelii Keywords: Plasmodium, CRISPR, CRISPRi, Ribozyme, HDR, ALBA bioRxiv preprint doi: https://doi.org/10.1101/481416; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Abstract 2 Functional characterization of genes in Plasmodium parasites often relies on genetic 3 manipulations to disrupt or modify a gene-of-interest. However, these approaches are limited by 4 the time required to generate transgenic parasites for P. falciparum and the availability of a 5 single drug selectable marker for P. yoelii. In both cases, there remains a risk of disrupting native 6 gene regulatory elements with the introduction of exogenous sequences. To address these 7 limitations, we have developed CRISPR-RGR, a SpCas9-based gene editing system for 8 Plasmodium that utilizes a Ribozyme-Guide-Ribozyme (RGR) sgRNA expression strategy. 9 Using this system with P. yoelii, we demonstrate that both gene disruptions and coding sequence 10 insertions are efficiently generated, producing marker-free and scar-free parasites with homology 11 arms as short as 80-100bp. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group. -

CRISPR/Cas9 with Single Guide RNA Expression Driven by Small Trna Promoters Showed Reduced Editing Efficiency Compared to a U6 Promoter
Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press LETTER TO THE EDITOR CRISPR/Cas9 with single guide RNA expression driven by small tRNA promoters showed reduced editing efficiency compared to a U6 promoter YUDA WEI,1,2 YAN QIU,1,2 YANHAO CHEN,1,2 GAIGAI LIU,1,2 YONGXIAN ZHANG,1,2 LUWEI XU,3 and QIURONG DING1,2 1CAS Key Laboratory of Nutrition and Metabolism, Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China 2University of Chinese Academy of Sciences, Shanghai 200031, China 3Taizhou Hospital of Traditional Chinese Medicine, Taizhou 225300, China ABSTRACT Multiplex genome engineering in vivo with CRISPR/Cas9 shows great promise as a potential therapeutic approach. The ability to incorporate multiple single guide RNA (sgRNA) cassettes together with Cas9 gene expression in one AAV vector could greatly enhance the efficiency. In a recent Method article, Mefferd and coworkers indicated that small tRNA promoters could be used to drive sgRNA expression to facilitate the construction of a more effective AAV vector. In contrast, we found that when targeting endogenous genomic loci, CRISPR/Cas9 with tRNA promoter-driven sgRNA expression showed much reduced genome editing activity, compared with significant cleavage with U6 promoter-driven sgRNA expression. Though the underlying mechanisms are still under investigation, our study suggests that the CRISPR/Cas9 system with tRNA promoter- driven sgRNA expression needs to be reevaluated before it can be used for therapeutic genome editing. Keywords: CRISPR/Cas9; genome editing; tRNA promoter The use of clustered regularly interspaced short palindromic have a DNA packaging limit of ∼5 kb, and the standard repeats (CRISPR)/CRISPR-associated (Cas) systems for in AAV–SaCRISPR system almost fills the cargo limitation, it vivo genome editing has emerged as a potential treatment would be helpful to further reduce the total length of the for human diseases (Cox et al.