Wo 2007/122369 A2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download The
PROBING THE INTERACTION OF ASPERGILLUS FUMIGATUS CONIDIA AND HUMAN AIRWAY EPITHELIAL CELLS BY TRANSCRIPTIONAL PROFILING IN BOTH SPECIES by POL GOMEZ B.Sc., The University of British Columbia, 2002 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Experimental Medicine) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2010 © Pol Gomez, 2010 ABSTRACT The cells of the airway epithelium play critical roles in host defense to inhaled irritants, and in asthma pathogenesis. These cells are constantly exposed to environmental factors, including the conidia of the ubiquitous mould Aspergillus fumigatus, which are small enough to reach the alveoli. A. fumigatus is associated with a spectrum of diseases ranging from asthma and allergic bronchopulmonary aspergillosis to aspergilloma and invasive aspergillosis. Airway epithelial cells have been shown to internalize A. fumigatus conidia in vitro, but the implications of this process for pathogenesis remain unclear. We have developed a cell culture model for this interaction using the human bronchial epithelium cell line 16HBE and a transgenic A. fumigatus strain expressing green fluorescent protein (GFP). Immunofluorescent staining and nystatin protection assays indicated that cells internalized upwards of 50% of bound conidia. Using fluorescence-activated cell sorting (FACS), cells directly interacting with conidia and cells not associated with any conidia were sorted into separate samples, with an overall accuracy of 75%. Genome-wide transcriptional profiling using microarrays revealed significant responses of 16HBE cells and conidia to each other. Significant changes in gene expression were identified between cells and conidia incubated alone versus together, as well as between GFP positive and negative sorted cells. -

Effet De La Cryptorchidie Sur Le Transcriptome Testiculaire Humain
MARIE EVE BERGERON EFFET DE LA CRYPTORCHIDIE SUR LE TRANSCRIPTOME TESTICULAIRE HUMAIN Mémoire présenté à la Faculté des études supérieures et postdoctorales de l’Université Laval dans le cadre du programme de maîtrise en Physiologie-Endocrinologie pour l’obtention du grade de Maître ès sciences (M.Sc.) DÉPARTEMENT D’OBSTÉTRIQUE ET DE GYNÉCOLOGIE FACULTÉ DE MÉDECINE UNIVERSITÉ LAVAL QUÉBEC 2012 © Marie Eve Bergeron, 2012 Résumé Les niveaux d’expression de nombreux gènes peuvent être affectés par l’environnement et mener au développement de la cryptorchidie. Cette malformation congénitale est la plus commune dont une des conséquences majeures est l’infertilité masculine due au testicule non-descendu, auquel un risque plus élevé de cancer testiculaire est associé. L’expression des ARN totaux isolés à partir de biopsies testiculaires ont été analysés par micropuces, puis par une analyse bio-informatique et une validation par RT-qPCR de plusieurs gènes sélectionnés. Ces analyses m’ont permis d’identifier plus de deux milles candidats montrant une expression différente entre des sujets cryptorchides et normaux. Certains de ces gènes sélectionnés peuvent être associés à la descente testiculaire, d’autres au cancer testiculaire ou encore aux divers types cellulaires retrouvés dans cet organe. Les différences dans le transcriptome dues à la cryptorchidie vont nous aider à comprendre la cause génétique de cette maladie. ii Abstract Expression level of numerous genes may be affected by environmental condition and lead to development of cryptorchidism. The most common congenital malformation in male is cryptorchidism. One major consequence of this anomaly is infertility due to undescended testis, to which an increased risk of testicular cancer is associated. -

A Dissertation Entitled the Androgen Receptor
A Dissertation entitled The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit Submitted to the Graduate Faculty as partial fulfillment of the requirements for the PhD Degree in Biomedical science Dr. Manohar Ratnam, Committee Chair Dr. Lirim Shemshedini, Committee Member Dr. Robert Trumbly, Committee Member Dr. Edwin Sanchez, Committee Member Dr. Beata Lecka -Czernik, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2011 Copyright 2011, Mesfin Gonit This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit As partial fulfillment of the requirements for the PhD Degree in Biomedical science The University of Toledo August 2011 Prostate cancer depends on the androgen receptor (AR) for growth and survival even in the absence of androgen. In the classical models of gene activation by AR, ligand activated AR signals through binding to the androgen response elements (AREs) in the target gene promoter/enhancer. In the present study the role of AREs in the androgen- independent transcriptional signaling was investigated using LP50 cells, derived from parental LNCaP cells through extended passage in vitro. LP50 cells reflected the signature gene overexpression profile of advanced clinical prostate tumors. The growth of LP50 cells was profoundly dependent on nuclear localized AR but was independent of androgen. Nevertheless, in these cells AR was unable to bind to AREs in the absence of androgen. -

Targeting EZH2 Increases Therapeutic Efficacy of PD-1 Check-Point Blockade in Models of Prostate Cancer Supplement Figures and T
Targeting EZH2 Increases Therapeutic Efficacy of PD-1 Check-Point Blockade in Models of Prostate Cancer Supplement Figures and Tables 1 Fig. S1. (A) Schema and genotyping PCR example for the creation of EM and EMC genetically engineered mice. (B) Three-dimensional PCa organoids generated from EM mice (without PSACreERT2) alleles. When treated with tamoxifen, demonstrates no loss of H3K27me3 or EDU staining, indicating specificity of tamoxifen-PSACreERT2 mediated deletion of the Ezh2 set domain. (C) Principle component analysis (PCA) following chemical and genetic inhibition of Ezh2 catalytic function results in significant changes in gene expression. 2 Fig. S2. (A) A 29-gene signature derived from Fig. 1C demonstrates complete independence from a previously published polycomb repression signature. (B) Our 29 gene signature demonstrates significant correlation with a previously published polycomb repression signature in 2 independent human PCa gene expression datasets. (C) EZH2 activity is not determined by EZH2 mRNA expression. 3 Fig. S3. A 29-gene signature derived from Fig 1C was used to generate signature scores for each patient within four independent human prostate cancer RNA-seq datasets. Patients were ranked highest score to lowest score and subject to quartile separation. First (blue) and fourth (red) quartiles were analyzed by supervised clustering to demonstrate expression differences within patients with most lowest EZH2 activity and most highest EZH2 activity. 4 Fig. S4. Genes representing IFN signaling (STAT1, IRF9), Th1 chemokines (CXCL10, CXCL11), and MHC Class I molecules (B2M, HLA-A) were shown to be enriched in PCa patients with low EZH2 activity. 5 Fig. S5. Treatment of 22Rv1 human 2D cell lines with the demonstrated conditions for 96 hours show that EZH2 inhibition increases expression of dsRNA (green = dsRNA, blue = nuclei). -

Supplementary Table 1. Differentially Expressed Transcripts Between Insulin Treated (T) and Insulin Deprived (D) Type 1 Diabetic Patients
Table Legends: Supplementary Table 1. Differentially expressed transcripts between insulin treated (T) and insulin deprived (D) type 1 diabetic patients. P values were calculated using paired t-tests, and not adjusted for multiple comparison errors. Annotations for each transcript were provided by Affymetrix NetAffx Analysis Center. Supplementary Table 2. Differentially expressed mitochondrial genes between insulin treated (T) and insulin deprived (D) type 1 diabetic patients. This EXCEL file has multiple data sheets. The data sheet “MITO GENE” lists the 68 mitochondrial genes, which were grouped into 11 functional groups in 11 data sheets. P values were calculated using paired t-tests, and not adjusted for multiple comparison errors. Annotations for each transcript were provided by Affymetrix NetAffx Analysis Center. OXPHOS- oxidative phosphorylation; TCA - tricarboxylic acid cycle. Supplementary Table 3. Differentially expressed pathways by Ingenuity Pathway Analysis (IPA). This EXCEL file has two data sheets that listed the significantly up- and down-regulated pathways in insulin-deprived vs. insulin- treated patients, respectively. P values associated with each pathway were calculated by IPA. Genes were represented by HUGO symbols. Supplementary Table 1 probe set T1 T2 T3 T4 T5 T6 T7 T8 214046_at 71.6231 78.4793 69.0236 71.6228 71.3996 61.9447 66.1693 61.9322 240081_at 145.816 133.058 155.18 126.734 137.149 130.863 157.818 127.17 227644_at 132.603 132.984 144.789 127.445 134.055 115.207 141.926 126.212 233926_at 13.1944 14.7052 12.8407 -
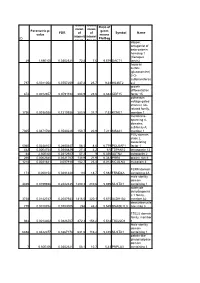
ID Parametric P
Geom Geom Ratio of mean mean Parametric p- geom FDR of of Symbol Name value means intensit intensi ID Pla/Bag ies in ties in dapper, antagonist of beta-catenin, homolog 1 (Xenopus 29 1,69E-05 0,0302321 72,8 7,6 9,579 DACT1 laevis) heparan sulfate (glucosamine) 3-O- sulfotransferas 797 0,0011054 0,0757259 237,2 25,7 9,23 HS3ST2 e 2 growth differentiation 874 0,0012657 0,0791194 204,9 23,6 8,682 GDF15 factor 15 potassium voltage-gated channel, Isk- related family, 1756 0,0038928 0,1210936 283,5 37,7 7,52 KCNE1 member 1 membrane- spanning 4- domains, subfamily A, 7305 0,0471766 0,3530226 150,7 20,9 7,211 MS4A1 member 1 POU domain, class 2, associating 5060 0,0248103 0,2680447 58,3 8,6 6,779 POU2AF1 factor 1 442 0,0004744 0,0586829 27,3 4,2 6,5 TSPAN12 tetraspanin 12 44 2,60E-05 0,0312927 57,2 9 6,356 ASTN2 astrotactin 2 266 0,0002545 0,0521767 138,9 21,9 6,342 PRR6 proline rich 6 1218 0,0021841 0,097918 152,7 25,4 6,012 MCOLN3 mucolipin 3 FERM domain 173 0,000154 0,0481428 110 18,7 5,882 FRMD4A containing 4A male sterility domain 4469 0,0199658 0,2442435 1200,3 210,6 5,699 MLSTD1 containing 1 aldehyde dehydrogenas e 1 family, 3738 0,0142747 0,2087933 1816,5 320,1 5,675 ALDH1A2 member A2 deoxyribonucle 779 0,0010754 0,0753005 268 48,3 5,549 DNASE1L3 ase I-like 3 TSC22 domain family, member 944 0,0014302 0,0826707 872,3 158,2 5,514 TSC22D1 1 Male sterility domain 6858 0,0422377 0,3367178 631,6 116,2 5,435 MLSTD1 containing 1 patatin-like phospholipase domain 7 6,60E-06 0,0302321 58,1 10,7 5,43 PNPLA3 containing 3 hypothetical protein 1857 -

Graded Co-Expression of Ion Channel, Neurofilament, and Synaptic Genes in Fast- Spiking Vestibular Nucleus Neurons
Research Articles: Cellular/Molecular Graded co-expression of ion channel, neurofilament, and synaptic genes in fast- spiking vestibular nucleus neurons https://doi.org/10.1523/JNEUROSCI.1500-19.2019 Cite as: J. Neurosci 2019; 10.1523/JNEUROSCI.1500-19.2019 Received: 26 June 2019 Revised: 11 October 2019 Accepted: 25 October 2019 This Early Release article has been peer-reviewed and accepted, but has not been through the composition and copyediting processes. The final version may differ slightly in style or formatting and will contain links to any extended data. Alerts: Sign up at www.jneurosci.org/alerts to receive customized email alerts when the fully formatted version of this article is published. Copyright © 2019 the authors 1 Graded co-expression of ion channel, neurofilament, and synaptic genes in fast-spiking 2 vestibular nucleus neurons 3 4 Abbreviated title: A fast-spiking gene module 5 6 Takashi Kodama1, 2, 3, Aryn Gittis, 3, 4, 5, Minyoung Shin2, Keith Kelleher2, 3, Kristine Kolkman3, 4, 7 Lauren McElvain3, 4, Minh Lam1, and Sascha du Lac1, 2, 3 8 9 1 Johns Hopkins University School of Medicine, Baltimore MD, 21205 10 2 Howard Hughes Medical Institute, La Jolla, CA, 92037 11 3 Salk Institute for Biological Studies, La Jolla, CA, 92037 12 4 Neurosciences Graduate Program, University of California San Diego, La Jolla, CA, 92037 13 5 Carnegie Mellon University, Pittsburgh, PA, 15213 14 15 Corresponding Authors: 16 Takashi Kodama ([email protected]) 17 Sascha du Lac ([email protected]) 18 Department of Otolaryngology-Head and Neck Surgery 19 The Johns Hopkins University School of Medicine 20 Ross Research Building 420, 720 Rutland Avenue, Baltimore, Maryland, 21205 21 22 23 Conflict of Interest 24 The authors declare no competing financial interests. -

Rabbit Anti-CLCA3 Antibody-SL13706R
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-CLCA3 antibody SL13706R Product Name: CLCA3 Chinese Name: 钙激活氯离子Channel protein3抗体 Calcium activated chloride channel 3; Calcium-activated chloride channel regulator family member 3; Chloride channel accessory 3 pseudogene; chloride channel calcium Alias: activated 3; Chloride channel calcium activated family member 3; Clca1; Clca2; CLCA3P; Gob 5; Gob-5; Gob5; hCLCA3; mCLCA3; MGC143984; MGC143985. Organism Species: Rabbit Clonality: Polyclonal React Species: Mouse,Rat,Dog, WB=1:500-2000ELISA=1:500-1000IHC-P=1:400-800IHC-F=1:400-800ICC=1:100- 500IF=1:100-500(Paraffin sections need antigen repair) Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 98kDa Cellular localization: cytoplasmic Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLHwww.sunlongbiotech.com conjugated synthetic peptide derived from mouse CLCA3:811-913/913 Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. PubMed: PubMed The calcium-activated chloride channel (CLCA) protein family, which includes the human homologs CLCA1 and CLCA2, display distinct tissue distribution patterns. -

Analysis of the Mouse Transcriptome for Genes Involved in the Function of the Nervous System Stefano Gustincich,1,13,14 Serge Batalov,2 Kirk W
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Analysis of the Mouse Transcriptome for Genes Involved in the Function of the Nervous System Stefano Gustincich,1,13,14 Serge Batalov,2 Kirk W. Beisel,3 Hidemasa Bono,4 Piero Carninci,4,5 Colin F. Fletcher,2,6 Sean Grimmond,7 Nobutaka Hirokawa,8 Erich D. Jarvis,9 Tim Jegla,2 Yuka Kawasawa,10 Julianna LeMieux,1 Harukata Miki,8 Elio Raviola,1 Rohan D. Teasdale,7 Naoko Tominaga,4 Ken Yagi,4 Andreas Zimmer,11 RIKEN GER Group4 and GSL Members,5,12 Yoshihide Hayashizaki,4,5 and Yasushi Okazaki4,5 1Department of Neurobiology, Harvard Medical School, Boston, Massachusetts 02115, USA; 2Genomics Institute of the Novartis Research Foundation (GNF), San Diego, California 92121, USA; 3Boys Town National Research Hospital, Omaha, Nebraska 68131, USA; 4Laboratory for Genome Exploration Research Group, RIKEN Genomic Sciences Center (GSC), RIKEN Yokohama Institute, Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan; 5Genome Science Laboratory, RIKEN, Hireosawa, Wako, Saitama, 351-0198, Japan; 6The Scripps Research Institute, La Jolla, California 92037, USA; 7Institute for Molecule Bioscience and ARC Special Research Centre for Functional and Applied Genomics, University of Queensland, Q4072, Australia; 8Graduate School of Medicine, University of Tokyo, Tokyo, 113-0033, Japan; 9Duke University Medical Center, Department of Neurobiology, Durham, North Carolina 27710, USA; 10Howard Hughes Medical Institute, Department of Molecular Genetics, -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

(12) United States Patent (10) Patent No.: US 7,666,610 B2 Saitoh Et Al
USOO766661 OB2 (12) United States Patent (10) Patent No.: US 7,666,610 B2 Saitoh et al. (45) Date of Patent: Feb. 23, 2010 (54) EXPRESSING TRANSPORTERS ON VIRAL JP 11-172 1, 1999 ENVELOPES JP 2001-197846 T 2001 JP 2001-1394.96 5, 2005 (75) Inventors: Ryoichi Saitoh, Shizuoka (JP); KR 99071666 9, 1999 Toshihiko Ohtomo, Shizuoka (JP); WO WO97, 19919 6, 1997 Masayuki Tsuchiya, Shizuoka (JP) WO WO 98.46777 10, 1998 WO WOOO.28O16 5, 2000 (73) Assignee: Chugai Seiyaku Kabushiki Kaisha, WO WOO3/033024 4/2003 Tokyo (JP) WO WO 03/0476.21 6, 2003 WO WO 03/08311.6 A1 10, 2003 (*) Notice: Subject to any disclaimer, the term of this WO WOO3/104453 12/2003 patent is extended or adjusted under 35 U.S.C. 154(b) by 283 days. OTHER PUBLICATIONS (21) Appl. No.: 10/509,343 Hsu et al., Overexpression of human intestinal oligopeptide trans porter in mammalian cells via adenoviral transduction, 1998, Phar (22) PCT Filed: Mar. 28, 2003 maceutical Research, vol. 15, No. 9, pp. 1376-1381.* Garcia et al., cDNA cloning of MCT2, a second monocarboxylate (86). PCT No.: PCT/UP03/03975 transporter expressed in different cells than MCT1, 1995, Journal of Biological Chemistry, vol. 270, No. 4, pp. 1843-1849.* S371 (c)(1), Miyasaka, et al., Characterization of human taurine transporter (2), (4) Date: Jun. 21, 2005 expressed in insect cells using recombinant baculovirus, 2001, Pro tein Expression and Purification, vol. 23, pp. 389-397.* (87) PCT Pub. No.: WO03/083116 Hsu et al., Overexpression of human intestinal oligopeptide trans porter in mammalian cells via adenoviral transduction, 1998, Phar PCT Pub. -

Supplementary Table 1
Supplementary Table 1. List of genes that encode proteins contianing cell surface epitopes and are represented on Agilent human 1A V2 microarray chip (2,177 genes) Agilent Probe ID Gene Symbol GenBank ID UniGene ID A_23_P103803 FCRH3 AF459027 Hs.292449 A_23_P104811 TREH AB000824 Hs.129712 A_23_P105100 IFITM2 X57351 Hs.174195 A_23_P107036 C17orf35 X51804 Hs.514009 A_23_P110736 C9 BC020721 Hs.1290 A_23_P111826 SPAM1 NM_003117 Hs.121494 A_23_P119533 EFNA2 AJ007292 No-Data A_23_P120105 KCNS3 BC004987 Hs.414489 A_23_P128195 HEM1 NM_005337 Hs.182014 A_23_P129332 PKD1L2 BC014157 Hs.413525 A_23_P130203 SYNGR2 AJ002308 Hs.464210 A_23_P132700 TDGF1 X14253 Hs.385870 A_23_P1331 COL13A1 NM_005203 Hs.211933 A_23_P138125 TOSO BC006401 Hs.58831 A_23_P142830 PLA2R1 U17033 Hs.410477 A_23_P146506 GOLPH2 AF236056 Hs.494337 A_23_P149569 MG29 No-Data No-Data A_23_P150590 SLC22A9 NM_080866 Hs.502772 A_23_P151166 MGC15619 BC009731 Hs.334637 A_23_P152620 TNFSF13 NM_172089 Hs.54673 A_23_P153986 KCNJ3 U39196 No-Data A_23_P154855 KCNE1 NM_000219 Hs.121495 A_23_P157380 KCTD7 AK056631 Hs.520914 A_23_P157428 SLC10A5 AK095808 Hs.531449 A_23_P160159 SLC2A5 BC001820 Hs.530003 A_23_P162162 KCTD14 NM_023930 Hs.17296 A_23_P162668 CPM BC022276 Hs.334873 A_23_P164341 VAMP2 AL050223 Hs.25348 A_23_P165394 SLC30A6 NM_017964 Hs.552598 A_23_P167276 PAQR3 AK055774 Hs.368305 A_23_P170636 KCNH8 AY053503 Hs.475656 A_23_P170736 MMP17 NM_016155 Hs.159581 A_23_P170959 LMLN NM_033029 Hs.518540 A_23_P19042 GABRB2 NM_021911 Hs.87083 A_23_P200361 CLCN6 X83378 Hs.193043 A_23_P200710 PIK3C2B