Role of the Visual Experience-Dependent Nascent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Ubiquitin Proteasome System Is Required for Cell Proliferation of The
RESEARCH ARTICLE 3257 Development 137, 3257-3268 (2010) doi:10.1242/dev.053124 © 2010. Published by The Company of Biologists Ltd The ubiquitin proteasome system is required for cell proliferation of the lens epithelium and for differentiation of lens fiber cells in zebrafish Fumiyasu Imai1, Asuka Yoshizawa1, Noriko Fujimori-Tonou2, Koichi Kawakami3 and Ichiro Masai1,* SUMMARY In the developing vertebrate lens, epithelial cells differentiate into fiber cells, which are elongated and flat in shape and form a multilayered lens fiber core. In this study, we identified the zebrafish volvox (vov) mutant, which shows defects in lens fiber differentiation. In the vov mutant, lens epithelial cells fail to proliferate properly. Furthermore, differentiating lens fiber cells do not fully elongate, and the shape and position of lens fiber nuclei are affected. We found that the vov mutant gene encodes Psmd6, the subunit of the 26S proteasome. The proteasome regulates diverse cellular functions by degrading polyubiquitylated proteins. Polyubiquitylated proteins accumulate in the vov mutant. Furthermore, polyubiquitylation is active in nuclei of differentiating lens fiber cells, suggesting roles of the proteasome in lens fiber differentiation. We found that an E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) is involved in lens defects in the vov mutant. These data suggest that the ubiquitin proteasome system is required for cell proliferation of lens epithelium and for the differentiation of lens fiber cells in zebrafish. KEY WORDS: Ubiquitin, Proteasome, Lens, Zebrafish, Psmd6, APC/C INTRODUCTION acid DNase (DLAD; Dnase2b). However, it is unclear how lens During development, the lens placode delaminates from the morphogenesis and organelle loss are related to these transcription epidermal ectoderm and forms the lens vesicle. -
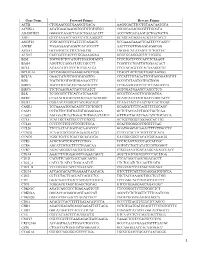
Gene Name Forward Primer Reverse Primer ACTB
Gene Name Forward Primer Reverse Primer ACTB CTGGAACGGTGAAGGTGACA AAGGGACTTCCTGTAACAATGCA ACVRL1 ACATGAAGAAGGTGGTGTGTGTGG CGGGCAGAGGGGTTTGGGTA ADAMDEC1 GGGGCCAGACTACACTGAAACATT ACCCGTCACAAGTACTGATGCTG AHI1 GTCCAAAACTACCCCATCAAGGCT GCAGCACAGGAACGTATCACCT ANGPT2 TGGCAGCGTTGATTTTCAGAGG GCGAAACAAACTCATTTCCCAGCC ANPEP TGAAGAAGCAGGTCACACCCCT AACTCCGTTGGAGCAGGCGG APOA1 GCCGTGCTCTTCCTGACGG TGGGACACATAGTCTCTGCCGC ATXN7 CACCGCCCACTCTGGAAAAGAA GGGTGCAGGGCTTCTTGGTG B2M TGCTGTCTCCATGTTTGATGTATCT TCTCTGCTCCCCACCTCTAAGT BAG4 AGGTTCCAGGATATCCGCCTT TCGGTCCTGATTGTGGAACACT BCL2 ACAACATCGCCCTGTGGATGA CCGTACAGTTCCACAAAGGCAT BCL2L14 GCTCAGGGTCAAAGGACGTTGG TCAGCTACTCGGTTGGCAATGG BCL7A GAACCATGTCGGGCAGGTCG CCCATTTGTAGATTCGTAGGGATGTGT BIN1 TGCTGTCGTGGTGGAGACCTTC GCCGTGTAGTCGTGCTGGG BIRC3 TGCTATCCACATCAGACAGCCC TCTGAATGGTCTTCTCCAGGTTCA BIRC5 TTCTCAAGGACCACCGCATCT AGTGGATGAAGCCAGCCTCG BLK TCGGGGTCTTCACCATCAAAGC GCGCTCCAGGTTGCGGATGA BTRC CCAAATGTGTCATTACCAACATGGGC GCAGCACATAGTGATTTGGCATCC BUB3 CGGAACATGGGTTACGTGCAGC CCAAATACTCAACTGCCACTCGGC CAGE1 TCCAAAATGCACAGTCTTCTGGCT GGAGGCTCTTCAGTTTTTGCAGC CASP1 CCTGTTCCTGTGATGTGGAGGAAA GCTCTACCATCTGGCTGCTCAA CASP3 AGCGAATCAATGGACTCTGGAATATCC GTTTGCTGCATCGACATCTGTACCA CCL5 TCATTGCTACTGCCCTCTGCG ACTGCTGGGTTGGAGCACTTG CCL18 CCCTCCTTGTCCTCGTCTGCA GCACTGGGGGCTGGTTTCAG CCL26 TTCCAATACAGCCACAAGCCCC GGATGGGTACAGACTTTCTTGCCTC CCND2 TCAAGTGCGTGCAGAAGGACAT CTTCGCACTTCTGTTCCTCACA CCND3 TGGCTGCTGTGATTGCACATGA GATGGCGGGTACATGGCAAAGG CCR3 ACGCTGCTCTGCTTCCTGG TCCTCAGTTCCCCACCATCGC CCR4 AGCATCGTGCTTCCTGAGCAA GGTGTCTGCTATATCCGTGGGGT CCR7 AGACAGGGGTAGTGCGAGGC -
![Myelogenous Leukemia 8;21 Translocation: Evidence That C-Mos Is Not Translocated [Somatic Cell Hybrids/C-Myc/Gene Mapping/Superoxide Dismutase (Soluble)] HARRY A](https://docslib.b-cdn.net/cover/6910/myelogenous-leukemia-8-21-translocation-evidence-that-c-mos-is-not-translocated-somatic-cell-hybrids-c-myc-gene-mapping-superoxide-dismutase-soluble-harry-a-2176910.webp)
Myelogenous Leukemia 8;21 Translocation: Evidence That C-Mos Is Not Translocated [Somatic Cell Hybrids/C-Myc/Gene Mapping/Superoxide Dismutase (Soluble)] HARRY A
Proc. Natl. Acad. Sci. USA Vol. 82, pp. 464-468, January 1985 Genetics Isolation and analysis of the 21q+ chromosome in the acute myelogenous leukemia 8;21 translocation: Evidence that c-mos is not translocated [somatic cell hybrids/c-myc/gene mapping/superoxide dismutase (soluble)] HARRY A. DRABKIN*t, MANUEL DIAZt, CYNTHIA M. BRADLEY*, MICHELLE M. LE BEAUS, JANET D. ROWLEYt, AND DAVID PATTERSON*§¶ *The Eleanor Roosevelt Institute for Cancer Research, 4200 East Ninth Avenue, B-129, and tThe Division of Medical Oncology and Departments of Medicine and ¶Biochemistry, Biophysics and Genetics, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Denver, CO 80262; and tSection of Hematology-Oncology, Department of Medicine, Pritzker School of Medicine, The University of Chicago, Chicago, IL 60637 Contributed by Janet D. Rowley, September 4, 1984 ABSTRACT Acute myelogenous leukemia (AML), sub- light chain genes are translocated to the 3' region of c-myc group M2, is associated with a nonrandom chromosomal (4-7). translocation, t(8;21)(q22,q22). The oncogene c-mos also has Somatic cell genetic approaches have been used also to been localized to the q22 band on chromosome 8. There is also show that, in Philadelphia (Ph')-positive chronic myeloge- evidence that genes on chromosome 21 may be important in nous leukemia, c-abl oncogene sequences are translocated the development of leukemia. To determine whether the c-mos from chromosome 9 to the Ph' (22q-) chromosome (8). oncogene has been translocated in AML-M2 with this translo- Analysis of cells from two Ph'-positive patients has revealed cation and to isolate DNA sequences and genes from these two that the breakpoint in chromosome 9 is near c-abl (9). -

Downregulation of Carnitine Acyl-Carnitine Translocase by Mirnas
Page 1 of 288 Diabetes 1 Downregulation of Carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion Mufaddal S. Soni1, Mary E. Rabaglia1, Sushant Bhatnagar1, Jin Shang2, Olga Ilkayeva3, Randall Mynatt4, Yun-Ping Zhou2, Eric E. Schadt6, Nancy A.Thornberry2, Deborah M. Muoio5, Mark P. Keller1 and Alan D. Attie1 From the 1Department of Biochemistry, University of Wisconsin, Madison, Wisconsin; 2Department of Metabolic Disorders-Diabetes, Merck Research Laboratories, Rahway, New Jersey; 3Sarah W. Stedman Nutrition and Metabolism Center, Duke Institute of Molecular Physiology, 5Departments of Medicine and Pharmacology and Cancer Biology, Durham, North Carolina. 4Pennington Biomedical Research Center, Louisiana State University system, Baton Rouge, Louisiana; 6Institute for Genomics and Multiscale Biology, Mount Sinai School of Medicine, New York, New York. Corresponding author Alan D. Attie, 543A Biochemistry Addition, 433 Babcock Drive, Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, (608) 262-1372 (Ph), (608) 263-9608 (fax), [email protected]. Running Title: Fatty acyl-carnitines enhance insulin secretion Abstract word count: 163 Main text Word count: 3960 Number of tables: 0 Number of figures: 5 Diabetes Publish Ahead of Print, published online June 26, 2014 Diabetes Page 2 of 288 2 ABSTRACT We previously demonstrated that micro-RNAs 132 and 212 are differentially upregulated in response to obesity in two mouse strains that differ in their susceptibility to obesity-induced diabetes. Here we show the overexpression of micro-RNAs 132 and 212 enhances insulin secretion (IS) in response to glucose and other secretagogues including non-fuel stimuli. We determined that carnitine acyl-carnitine translocase (CACT, Slc25a20) is a direct target of these miRNAs. -

Gene Mapping and Medical Genetics Human Chromosome 8
J Med Genet: first published as 10.1136/jmg.25.11.721 on 1 November 1988. Downloaded from Gene mapping and medical genetics Journal of Medical Genetics 1988, 25, 721-731 Human chromosome 8 STEPHEN WOOD From the Department of Medical Genetics, University of British Columbia, 6174 University Boulevard, Vancouver, British Columbia, Canada V6T IWS. SUMMARY The role of human chromosome 8 in genetic disease together with the current status of the genetic linkage map for this chromosome is reviewed. Both hereditary genetic disease attributed to mutant alleles at gene loci on chromosome 8 and neoplastic disease owing to somatic mutation, particularly chromosomal translocations, are discussed. Human chromosome 8 is perhaps best known for its In an era when complete sequencing of the human involvement in Burkitt's lymphoma and as the genome is being proposed, it is appropriate for location of the tissue plasminogen activator gene, medical geneticists to accept the challenge of defining by copyright. PLAT, which has been genetically engineered to the set of loci that have mutant alleles causing provide a natural fibrinolytic product for emergency hereditary disease. The fundamental genetic tool of use in cardiac disease. Since chromosome 8 repre- linkage mapping can now be applied, owing largely sents about 5% of the human genome, we may to progress in defining RFLP markers.3 4 This expect it to carry about 5% of human gene loci. This review will focus on genetic disease associated with would correspond to about 90 of the fully validated chromosome 8 loci and the status ofthe chromosome 8 phenotypes in the MIM7 catalogue.' The 27 genes linkage map. -

SF3B1-Mutated Chronic Lymphocytic Leukemia Shows Evidence Of
SF3B1-mutated chronic lymphocytic leukemia shows evidence of NOTCH1 pathway activation including CD20 downregulation by Federico Pozzo, Tamara Bittolo, Erika Tissino, Filippo Vit, Elena Vendramini, Luca Laurenti, Giovanni D'Arena, Jacopo Olivieri, Gabriele Pozzato, Francesco Zaja, Annalisa Chiarenza, Francesco Di Raimondo, Antonella Zucchetto, Riccardo Bomben, Francesca Maria Rossi, Giovanni Del Poeta, Michele Dal Bo, and Valter Gattei Haematologica 2020 [Epub ahead of print] Citation: Federico Pozzo, Tamara Bittolo, Erika Tissino, Filippo Vit, Elena Vendramini, Luca Laurenti, Giovanni D'Arena, Jacopo Olivieri, Gabriele Pozzato, Francesco Zaja, Annalisa Chiarenza, Francesco Di Raimondo, Antonella Zucchetto, Riccardo Bomben, Francesca Maria Rossi, Giovanni Del Poeta, Michele Dal Bo, and Valter Gattei SF3B1-mutated chronic lymphocytic leukemia shows evidence of NOTCH1 pathway activation including CD20 downregulation. Haematologica. 2020; 105:xxx doi:10.3324/haematol.2020.261891 Publisher's Disclaimer. E-publishing ahead of print is increasingly important for the rapid dissemination of science. Haematologica is, therefore, E-publishing PDF files of an early version of manuscripts that have completed a regular peer review and have been accepted for publication. E-publishing of this PDF file has been approved by the authors. After having E-published Ahead of Print, manuscripts will then undergo technical and English editing, typesetting, proof correction and be presented for the authors' final approval; the final version of the manuscript will -

Koch Shrna Gene Webpage
Symbol SEPT9 ADAM30 AEN AMBP ARHGEF12 ATG16L2 BCAS3 A1CF ADAM32 AFF3 AMBRA1 ARHGEF17 ATG2A BCKDK AAK1 ADAM33 AGAP2 AMHR2 ARHGEF2 ATG3 BCL10 AATK ADAM7 AGER AMPH ARHGEF4 ATG4B BCL11A ABCA1 ADAM8 AGK ANAPC2 ARHGEF6 ATG4C BCL11B ABCA3 ADAM9 AGL ANG ARHGEF7 ATG4D BCL2 ABCB1 ADAMDEC1 AGPAT9 ANGPT2 ARID1A ATG5 BCL2L1 ABCB4 ADAMTS1 AGR3 ANGPTL4 ARID1B ATG7 BCL2L11 ABCC1 ADAMTS10 AHR ANKK1 ARID2 ATM BCL2L2 ABCC10 ADAMTS12 AIMP2 ANKRD30A ARID3A ATMIN BCL3 ABCC2 ADAMTS13 AIP ANO1 ARID3B ATP1B3 BCL6 ABCG2 ADAMTS14 AJAP1 ANXA1 ARID4B ATP2B4 BCL7A ABI1 ADAMTS15 AK1 ANXA2 ARID5A ATP7A BCL9 ABL1 ADAMTS16 AK2 ANXA6 ARID5B ATP7B BCR ABL2 ADAMTS17 AK3 ANXA7 ARL11 ATR BECN1 ACIN1 ADAMTS18 AK4 APAF1 ARNT ATRX BFAR ACP1 ADAMTS19 AK5 APC ARSB ATXN1 BIK ACPP ADAMTS2 AK7 APCDD1 ARSG ATXN2 BIN1 ACSL4 ADAMTS20 AK8 APEX1 ASAP1 AURKA BIN2 ACTN1 ADAMTS3 AKAP1 APOBEC1 ASAP3 AURKB BIRC2 ACVR1 ADAMTS4 AKAP13 APOBEC2 ASB15 AURKC BIRC3 ACVR1B ADAMTS5 AKAP3 APOBEC3G ASCC1 AXIN1 BIRC5 ACVR1C ADAMTS7 AKAP8L AQP1 ASCC3 AXIN2 BIRC7 ACVR2A ADAMTS8 AKR1B10 AQP5 ASCL1 AXL BLCAP ACVR2B ADAMTS9 AKR1C1 AQP7 ASCL2 AZGP1 BLK ACVRL1 ADAR AKR1C3 AR ASF1A BACE1 BLM AD026 ADARB1 AKT1 ARAF ASH1L BAD BMI1 ADAM10 ADARB2 AKT2 AREG ASH2L BAG1 BMP2 ADAM11 ADAT2 AKT3 ARF1 ASNS BAG4 BMP2K ADAM12 ADCK1 ALCAM ARF4 ASPH BANF1 BMP2KL ADAM15 ADCK2 ALDH18A1 ARF5 ASPSCR1 BAP1 BMPR1A ADAM17 ADCK3 ALK ARF6 ASS1 BARD1 BMPR1B ADAM18 ADCK4 ALKBH2 ARHGAP12 ASTE1 BAX BMPR2 ADAM19 ADCK5 ALKBH3 ARHGAP22 ASXL1 BAZ1A BMX ADAM2 ADCY6 ALKBH8 ARHGAP25 ATF1 BAZ1B BNIP3 ADAM20 ADK ALOX15 ARHGAP26 ATF2 BAZ2A BPTF ADAM21 -

Genetic Technologies for Building a Better Mouse
Downloaded from genesdev.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW A most formidable arsenal: genetic technologies for building a better mouse James F. Clark,1 Colin J. Dinsmore,1 and Philippe Soriano Department of Cell, Developmental, and Regenerative Biology, Icahn School of Medicine at Mt. Sinai, New York, New York 10029, USA The mouse is one of the most widely used model organ- scribing Mendelian inheritance for mouse coat color char- isms for genetic study. The tools available to alter the acteristics (Cuénot 1902). Mendel himself had started mouse genome have developed over the preceding decades down this same road fifty years earlier, only for his bishop from forward screens to gene targeting in stem cells to the to snuff out the nascent breeding program because a monk recent influx of CRISPR approaches. In this review, we had no business, frankly, breeding (Paigen 2003). first consider the history of mice in genetic study, the Following Cuénot’s papers (Cuénot 1902), genetic re- development of classic approaches to genome modifica- search using the mouse began in earnest, often through tion, and how such approaches have been used and im- the study of spontaneous mouse mutants. One line of re- proved in recent years. We then turn to the recent surge search further explored the complicated genetics of pig- of nuclease-mediated techniques and how they are chang- mentation and the sometimes-surprising coincident ing the field of mouse genetics. Finally, we survey com- phenotypes. The Dominant white spotting allele W (an al- mon classes of alleles used in mice and discuss how lele of the receptor tyrosine kinase Kit) was found to affect they might be engineered using different methods. -

Supplemental Solier
Supplementary Figure 1. Importance of Exon numbers for transcript downregulation by CPT Numbers of down-regulated genes for four groups of comparable size genes, differing only by the number of exons. Supplementary Figure 2. CPT up-regulates the p53 signaling pathway genes A, List of the GO categories for the up-regulated genes in CPT-treated HCT116 cells (p<0.05). In bold: GO category also present for the genes that are up-regulated in CPT- treated MCF7 cells. B, List of the up-regulated genes in both CPT-treated HCT116 cells and CPT-treated MCF7 cells (CPT 4 h). C, RT-PCR showing the effect of CPT on JUN and H2AFJ transcripts. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. Supplementary Figure 3. Down-regulation of RNA degradation-related genes after CPT treatment A, “RNA degradation” pathway from KEGG. The genes with “red stars” were down- regulated genes after CPT treatment. B, Affy Exon array data for the “CNOT” genes. The log2 difference for the “CNOT” genes expression depending on CPT treatment was normalized to the untreated controls. C, RT-PCR showing the effect of CPT on “CNOT” genes down-regulation. HCT116 cells were treated with CPT (10 µM, 20 h) and CNOT6L, CNOT2, CNOT4 and CNOT6 mRNA were analysed by RT-PCR. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. D, CNOT6L down-regulation after CPT treatment. CNOT6L transcript was analysed by Q- PCR. Supplementary Figure 4. Down-regulation of ubiquitin-related genes after CPT treatment A, “Ubiquitin-mediated proteolysis” pathway from KEGG. -

Genetic Dissection of Susceptibility to Murine Ovarian Teratomas That Originate from Parthenogenetic Oocytes1
ICANCER RESEARCH 57. 590-593. February 15. 1997] Advances in Brief Genetic Dissection of Susceptibility to Murine Ovarian Teratomas That Originate from Parthenogenetic Oocytes1 Gang-Hong Lee,2 James M. Bugni, Masahiko Obata, Hiroyuki Nishimori, Katsuhiro Ogawa, and Norman R. Drinkwater Department of Pathology, Asahikawa Medical College, 4-5-3-11 Nishikagura. Asahikawa 078, Japan [G-H. L, M. 0., H. N.. K. 0.], and McArdle Laboratory for Cancer Research, University of Wisconsin Medical School, Madison, Wisconsin 53706 f.i. M. B., N. R. D.J Abstract Detectionof Ovarian Teratomas.Approximately100daysafterbirth, female mice were sacrificed by cervical dislocation, and ovarian tissues were The LTISv mousestrainis characterizedbyits abnormallyhighmci collected and fixed in formalin. The fixed ovarian tissues were examined dence of spontaneousovarian teratomas. These tumors have been shown histologically for teratomas using the diagnostic criteria reported by Stevens to originatefromparthenogeneticoocytes,whicharespontaneouslyin andVarnum(1).Foreachfemale(C57BL/6JX LT/Sv)F2animal,spleentissue duced to divide. Both spontaneous parthenogenesis and ovarian teratomas wasalsocollectedandfrozenasa sourceof genomicDNA. are extremelyrare for other mousestrains,includingC57BLI6J. To Genotypingfor SSRMarkers. Thegenotypesforthepolymorphicsimple identify the genes responsible for this unique phenotype of female LT/Sv sequence repeat marker loci (6, 7) were determined using PCR as described mice, we performed linkage analysis of female (C57BL/6J x LT/Sv)F2 -

Analyses of Compact Trichinella Kinomes Reveal a MOS-Like Protein Kinase with a Unique N-Terminal Domain
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors INVESTIGATION Analyses of Compact Trichinella Kinomes Reveal a MOS-Like Protein Kinase with a Unique N-Terminal Domain Andreas J. Stroehlein,* Neil D. Young,* Pasi K. Korhonen,* Bill C. H. Chang,*,† Paul W. Sternberg,‡,§ Giuseppe La Rosa,** Edoardo Pozio,** and Robin B. Gasser*,1 *Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, Parkville, Victoria 3010, Australia, †Yourgene Bioscience, Shu-Lin District, New Taipei City 23863, Taiwan, ‡Howard Hughes Medical Institute, Chevy Chase, § Maryland 20815-6789, Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California 91125, and **Istituto Superiore di Sanità, 00161 Rome, Italy ABSTRACT Parasitic worms of the genus Trichinella (phylum Nematoda; class Enoplea) represent a com- KEYWORDS plex of at least twelve taxa that infect a range of different host animals, including humans, around the world. parasitic worms They are foodborne, intracellular nematodes, and their life cycles differ substantially from those of other Trichinella nematodes. The recent characterization of the genomes and transcriptomes of all twelve recognized taxa of kinome Trichinella now allows, for the first time, detailed studies of their molecular biology. In the present study, we protein kinases defined, curated, and compared the protein kinase complements (kinomes) of Trichinella spiralis and T. protein pseudospiralis using an integrated bioinformatic workflow employing transcriptomic and genomic data sets. annotation We examined how variation in the kinome might link to unique aspects of Trichinella morphology, biology, and evolution. Furthermore, we utilized in silico structural modeling to discover and characterize a novel, MOS-like kinase with an unusual, previously undescribed N-terminal domain. -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein