The Ubiquitin Proteasome System Is Required for Cell Proliferation of The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Gene Clusters in Global Pathogenesis of Clear Cell Carcinoma of the Ovary Discovered by Integrated Analysis of Transcriptomes
International Journal of Environmental Research and Public Health Article Functional Gene Clusters in Global Pathogenesis of Clear Cell Carcinoma of the Ovary Discovered by Integrated Analysis of Transcriptomes Yueh-Han Hsu 1,2, Peng-Hui Wang 1,2,3,4,5 and Chia-Ming Chang 1,2,* 1 Department of Obstetrics and Gynecology, Taipei Veterans General Hospital, Taipei 112, Taiwan; [email protected] (Y.-H.H.); [email protected] (P.-H.W.) 2 School of Medicine, National Yang-Ming University, Taipei 112, Taiwan 3 Institute of Clinical Medicine, National Yang-Ming University, Taipei 112, Taiwan 4 Department of Medical Research, China Medical University Hospital, Taichung 440, Taiwan 5 Female Cancer Foundation, Taipei 104, Taiwan * Correspondence: [email protected]; Tel.: +886-2-2875-7826; Fax: +886-2-5570-2788 Received: 27 April 2020; Accepted: 31 May 2020; Published: 2 June 2020 Abstract: Clear cell carcinoma of the ovary (ovarian clear cell carcinoma (OCCC)) is one epithelial ovarian carcinoma that is known to have a poor prognosis and a tendency for being refractory to treatment due to unclear pathogenesis. Published investigations of OCCC have mainly focused only on individual genes and lack of systematic integrated research to analyze the pathogenesis of OCCC in a genome-wide perspective. Thus, we conducted an integrated analysis using transcriptome datasets from a public domain database to determine genes that may be implicated in the pathogenesis involved in OCCC carcinogenesis. We used the data obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) DataSets. We found six interactive functional gene clusters in the pathogenesis network of OCCC, including ribosomal protein, eukaryotic translation initiation factors, lactate, prostaglandin, proteasome, and insulin-like growth factor. -

Genome-Wide Transcript and Protein Analysis Reveals Distinct Features of Aging in the Mouse Heart
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.28.272260; this version posted April 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Genome-wide transcript and protein analysis reveals distinct features of aging in the mouse heart Isabela Gerdes Gyuricza1, Joel M. Chick2, Gregory R. Keele1, Andrew G. Deighan1, Steven C. Munger1, Ron Korstanje1, Steven P. Gygi3, Gary A. Churchill1 1The Jackson Laboratory, Bar Harbor, Maine 04609 USA; 2Vividion Therapeutics, San Diego, California 92121, USA; 3Harvard Medical School, Boston, Massachusetts 02115, USA Corresponding author: [email protected] Key words for online indexing: Heart Aging Transcriptomics Proteomics eQTL pQTL Stoichiometry ABSTRACT Investigation of the molecular mechanisms of aging in the human heart is challenging due to confounding factors, such as diet and medications, as well limited access to tissues. The laboratory mouse provides an ideal model to study aging in healthy individuals in a controlled environment. However, previous mouse studies have examined only a narrow range of the genetic variation that shapes individual differences during aging. Here, we analyzed transcriptome and proteome data from hearts of genetically diverse mice at ages 6, 12 and 18 months to characterize molecular changes that occur in the aging heart. Transcripts and proteins reveal distinct biological processes that are altered through the course of natural aging. Transcriptome analysis reveals a scenario of cardiac hypertrophy, fibrosis, and reemergence of fetal gene expression patterns. -

Dynamics of Protein Ubiquitination Upon Proteasome Modulation Karen Alexandra Sap
Dynamics of Protein Ubiquitination Upon Proteasome Modulation Karen Alexandra Sap Dynamics of Protein Ubiquitination Upon Proteasome Modulation A Quantitative Mass Spectrometry Approach Karen Alexandra Sap Dynamics of Protein Ubiquitination upon Proteasome Modulation A Quantitative Mass Spectrometry Approach Karen Alexandra Sap © 2018 Karen Sap Cover design: Karen Sap & Frank Sap ISBN: 978-94-93019-55-3 Printed by: ProefschriftMaken.nl || www.proefschriftmaken.nl Published by: ProefschriftMaken.nl || www.proefschriftmaken.nl The studies described in this thesis were performed in the Proteomics Center which is embedded in the Department of Biochemistry of the Erasmus University Medical Center in Rotterdam, The Netherlands The research described in this thesis was financially supported by the Netherlands Proteomics Center (project number 184.032.201) Dynamics of Protein Ubiquitination upon Proteasome Modulation A Quantitative Mass Spectrometry Approach Dynamiek van eiwit ubiquitinatie als gevolg van proteasoom modulatie Onderzocht door middel van kwantitatieve massaspectrometrie Proefschrift ter verkrijging van de graad van doctor aan de Erasmus Universiteit Rotterdam op gezag van de rector magnificus Prof.dr. R.C.M.E. Engels en volgens besluit van het College voor Promoties. De openbare verdediging zal plaatsvinden op donderdag 20 september 2018 om 11.30 uur door Karen Alexandra Sap geboren te Rotterdam Promotiecommissie Promotor: Prof.dr. C.P. Verrijzer Overige leden: Prof.dr. J.H. Gribnau Dr. J.A.F Marteijn Dr. A.C.O Vertegaal Copromotor: -

Arsenic Hexoxide Has Differential Effects on Cell Proliferation And
www.nature.com/scientificreports OPEN Arsenic hexoxide has diferential efects on cell proliferation and genome‑wide gene expression in human primary mammary epithelial and MCF7 cells Donguk Kim1,7, Na Yeon Park2,7, Keunsoo Kang3, Stuart K. Calderwood4, Dong‑Hyung Cho2, Ill Ju Bae5* & Heeyoun Bunch1,6* Arsenic is reportedly a biphasic inorganic compound for its toxicity and anticancer efects in humans. Recent studies have shown that certain arsenic compounds including arsenic hexoxide (AS4O6; hereafter, AS6) induce programmed cell death and cell cycle arrest in human cancer cells and murine cancer models. However, the mechanisms by which AS6 suppresses cancer cells are incompletely understood. In this study, we report the mechanisms of AS6 through transcriptome analyses. In particular, the cytotoxicity and global gene expression regulation by AS6 were compared in human normal and cancer breast epithelial cells. Using RNA‑sequencing and bioinformatics analyses, diferentially expressed genes in signifcantly afected biological pathways in these cell types were validated by real‑time quantitative polymerase chain reaction and immunoblotting assays. Our data show markedly diferential efects of AS6 on cytotoxicity and gene expression in human mammary epithelial normal cells (HUMEC) and Michigan Cancer Foundation 7 (MCF7), a human mammary epithelial cancer cell line. AS6 selectively arrests cell growth and induces cell death in MCF7 cells without afecting the growth of HUMEC in a dose‑dependent manner. AS6 alters the transcription of a large number of genes in MCF7 cells, but much fewer genes in HUMEC. Importantly, we found that the cell proliferation, cell cycle, and DNA repair pathways are signifcantly suppressed whereas cellular stress response and apoptotic pathways increase in AS6‑treated MCF7 cells. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

A Genome-Wide Dsrna Library Screen for Drosophila Genes That Regulate
Sung and Shears BMC Res Notes (2018) 11:884 https://doi.org/10.1186/s13104-018-3996-z BMC Research Notes RESEARCH NOTE Open Access A genome‑wide dsRNA library screen for Drosophila genes that regulate the GBP/ phospholipase C signaling axis that links infammation to aging Eui Jae Sung and Stephen B. Shears* Abstract Objective: Invertebrates are productive models for understanding how infammation, metabolism and aging are intertwined. We have deployed a dsRNA library screen to search for genes in Drosophila melanogaster—and hence 2 identify human orthologs—that encode participants in a G-protein coupled, Ca +-signaling pathway that regulates infammation, metabolism and lifespan. 2 Results: We analyzed receptor-dependent, phospholipase C/Ca + signaling responses to the growth-blocking peptide (GBP) cytokine in Drosophila S3 cells plated in 384-well plates containing dsRNAs that target approximately 14,000 Drosophila genes. We used Z-scores of < 3 or > 3 to defne gene hits. Filtering of ‘housekeeping’ genes − + 2 from these hits yielded a total of 82 and 61 Drosophila genes that either down-regulate or up-regulate Ca +-signaling, respectively; representatives from these two groups were validated. Human orthologs of our hits may be modulators 2 of Ca + signaling in general, as well as being candidates for acting in molecular pathways that interconnect aging and infammation. Keywords: Cytokine, Infammation, Metabolism, Calcium-signaling, G-proteins, Receptor Introduction Invertebrates are productive, genetically-tractable A systems-level understanding of cytokine-mediated, models for understanding how infammation and aging inter-tissue signaling can help to generate fundamen- are inter-related in humans [1, 4]. -

Gene Expression Profiles Reveal Alternative Targets of Therapeutic Intervention for the Treatment of Drug-Resistant Non-Small Cell Lung Cancers
University of Kentucky UKnowledge Theses and Dissertations--Pharmacy College of Pharmacy 2017 GENE EXPRESSION PROFILES REVEAL ALTERNATIVE TARGETS OF THERAPEUTIC INTERVENTION FOR THE TREATMENT OF DRUG-RESISTANT NON-SMALL CELL LUNG CANCERS Madeline J. Krentz Gober University of Kentucky, [email protected] Author ORCID Identifier: https://orcid.org/0000-0001-7761-6741 Digital Object Identifier: https://doi.org/10.13023/ETD.2017.309 Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Krentz Gober, Madeline J., "GENE EXPRESSION PROFILES REVEAL ALTERNATIVE TARGETS OF THERAPEUTIC INTERVENTION FOR THE TREATMENT OF DRUG-RESISTANT NON-SMALL CELL LUNG CANCERS" (2017). Theses and Dissertations--Pharmacy. 78. https://uknowledge.uky.edu/pharmacy_etds/78 This Doctoral Dissertation is brought to you for free and open access by the College of Pharmacy at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Pharmacy by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -
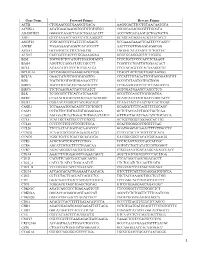
Gene Name Forward Primer Reverse Primer ACTB
Gene Name Forward Primer Reverse Primer ACTB CTGGAACGGTGAAGGTGACA AAGGGACTTCCTGTAACAATGCA ACVRL1 ACATGAAGAAGGTGGTGTGTGTGG CGGGCAGAGGGGTTTGGGTA ADAMDEC1 GGGGCCAGACTACACTGAAACATT ACCCGTCACAAGTACTGATGCTG AHI1 GTCCAAAACTACCCCATCAAGGCT GCAGCACAGGAACGTATCACCT ANGPT2 TGGCAGCGTTGATTTTCAGAGG GCGAAACAAACTCATTTCCCAGCC ANPEP TGAAGAAGCAGGTCACACCCCT AACTCCGTTGGAGCAGGCGG APOA1 GCCGTGCTCTTCCTGACGG TGGGACACATAGTCTCTGCCGC ATXN7 CACCGCCCACTCTGGAAAAGAA GGGTGCAGGGCTTCTTGGTG B2M TGCTGTCTCCATGTTTGATGTATCT TCTCTGCTCCCCACCTCTAAGT BAG4 AGGTTCCAGGATATCCGCCTT TCGGTCCTGATTGTGGAACACT BCL2 ACAACATCGCCCTGTGGATGA CCGTACAGTTCCACAAAGGCAT BCL2L14 GCTCAGGGTCAAAGGACGTTGG TCAGCTACTCGGTTGGCAATGG BCL7A GAACCATGTCGGGCAGGTCG CCCATTTGTAGATTCGTAGGGATGTGT BIN1 TGCTGTCGTGGTGGAGACCTTC GCCGTGTAGTCGTGCTGGG BIRC3 TGCTATCCACATCAGACAGCCC TCTGAATGGTCTTCTCCAGGTTCA BIRC5 TTCTCAAGGACCACCGCATCT AGTGGATGAAGCCAGCCTCG BLK TCGGGGTCTTCACCATCAAAGC GCGCTCCAGGTTGCGGATGA BTRC CCAAATGTGTCATTACCAACATGGGC GCAGCACATAGTGATTTGGCATCC BUB3 CGGAACATGGGTTACGTGCAGC CCAAATACTCAACTGCCACTCGGC CAGE1 TCCAAAATGCACAGTCTTCTGGCT GGAGGCTCTTCAGTTTTTGCAGC CASP1 CCTGTTCCTGTGATGTGGAGGAAA GCTCTACCATCTGGCTGCTCAA CASP3 AGCGAATCAATGGACTCTGGAATATCC GTTTGCTGCATCGACATCTGTACCA CCL5 TCATTGCTACTGCCCTCTGCG ACTGCTGGGTTGGAGCACTTG CCL18 CCCTCCTTGTCCTCGTCTGCA GCACTGGGGGCTGGTTTCAG CCL26 TTCCAATACAGCCACAAGCCCC GGATGGGTACAGACTTTCTTGCCTC CCND2 TCAAGTGCGTGCAGAAGGACAT CTTCGCACTTCTGTTCCTCACA CCND3 TGGCTGCTGTGATTGCACATGA GATGGCGGGTACATGGCAAAGG CCR3 ACGCTGCTCTGCTTCCTGG TCCTCAGTTCCCCACCATCGC CCR4 AGCATCGTGCTTCCTGAGCAA GGTGTCTGCTATATCCGTGGGGT CCR7 AGACAGGGGTAGTGCGAGGC -
![Myelogenous Leukemia 8;21 Translocation: Evidence That C-Mos Is Not Translocated [Somatic Cell Hybrids/C-Myc/Gene Mapping/Superoxide Dismutase (Soluble)] HARRY A](https://docslib.b-cdn.net/cover/6910/myelogenous-leukemia-8-21-translocation-evidence-that-c-mos-is-not-translocated-somatic-cell-hybrids-c-myc-gene-mapping-superoxide-dismutase-soluble-harry-a-2176910.webp)
Myelogenous Leukemia 8;21 Translocation: Evidence That C-Mos Is Not Translocated [Somatic Cell Hybrids/C-Myc/Gene Mapping/Superoxide Dismutase (Soluble)] HARRY A
Proc. Natl. Acad. Sci. USA Vol. 82, pp. 464-468, January 1985 Genetics Isolation and analysis of the 21q+ chromosome in the acute myelogenous leukemia 8;21 translocation: Evidence that c-mos is not translocated [somatic cell hybrids/c-myc/gene mapping/superoxide dismutase (soluble)] HARRY A. DRABKIN*t, MANUEL DIAZt, CYNTHIA M. BRADLEY*, MICHELLE M. LE BEAUS, JANET D. ROWLEYt, AND DAVID PATTERSON*§¶ *The Eleanor Roosevelt Institute for Cancer Research, 4200 East Ninth Avenue, B-129, and tThe Division of Medical Oncology and Departments of Medicine and ¶Biochemistry, Biophysics and Genetics, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Denver, CO 80262; and tSection of Hematology-Oncology, Department of Medicine, Pritzker School of Medicine, The University of Chicago, Chicago, IL 60637 Contributed by Janet D. Rowley, September 4, 1984 ABSTRACT Acute myelogenous leukemia (AML), sub- light chain genes are translocated to the 3' region of c-myc group M2, is associated with a nonrandom chromosomal (4-7). translocation, t(8;21)(q22,q22). The oncogene c-mos also has Somatic cell genetic approaches have been used also to been localized to the q22 band on chromosome 8. There is also show that, in Philadelphia (Ph')-positive chronic myeloge- evidence that genes on chromosome 21 may be important in nous leukemia, c-abl oncogene sequences are translocated the development of leukemia. To determine whether the c-mos from chromosome 9 to the Ph' (22q-) chromosome (8). oncogene has been translocated in AML-M2 with this translo- Analysis of cells from two Ph'-positive patients has revealed cation and to isolate DNA sequences and genes from these two that the breakpoint in chromosome 9 is near c-abl (9). -

Downregulation of Carnitine Acyl-Carnitine Translocase by Mirnas
Page 1 of 288 Diabetes 1 Downregulation of Carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion Mufaddal S. Soni1, Mary E. Rabaglia1, Sushant Bhatnagar1, Jin Shang2, Olga Ilkayeva3, Randall Mynatt4, Yun-Ping Zhou2, Eric E. Schadt6, Nancy A.Thornberry2, Deborah M. Muoio5, Mark P. Keller1 and Alan D. Attie1 From the 1Department of Biochemistry, University of Wisconsin, Madison, Wisconsin; 2Department of Metabolic Disorders-Diabetes, Merck Research Laboratories, Rahway, New Jersey; 3Sarah W. Stedman Nutrition and Metabolism Center, Duke Institute of Molecular Physiology, 5Departments of Medicine and Pharmacology and Cancer Biology, Durham, North Carolina. 4Pennington Biomedical Research Center, Louisiana State University system, Baton Rouge, Louisiana; 6Institute for Genomics and Multiscale Biology, Mount Sinai School of Medicine, New York, New York. Corresponding author Alan D. Attie, 543A Biochemistry Addition, 433 Babcock Drive, Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, (608) 262-1372 (Ph), (608) 263-9608 (fax), [email protected]. Running Title: Fatty acyl-carnitines enhance insulin secretion Abstract word count: 163 Main text Word count: 3960 Number of tables: 0 Number of figures: 5 Diabetes Publish Ahead of Print, published online June 26, 2014 Diabetes Page 2 of 288 2 ABSTRACT We previously demonstrated that micro-RNAs 132 and 212 are differentially upregulated in response to obesity in two mouse strains that differ in their susceptibility to obesity-induced diabetes. Here we show the overexpression of micro-RNAs 132 and 212 enhances insulin secretion (IS) in response to glucose and other secretagogues including non-fuel stimuli. We determined that carnitine acyl-carnitine translocase (CACT, Slc25a20) is a direct target of these miRNAs. -

Gene Mapping and Medical Genetics Human Chromosome 8
J Med Genet: first published as 10.1136/jmg.25.11.721 on 1 November 1988. Downloaded from Gene mapping and medical genetics Journal of Medical Genetics 1988, 25, 721-731 Human chromosome 8 STEPHEN WOOD From the Department of Medical Genetics, University of British Columbia, 6174 University Boulevard, Vancouver, British Columbia, Canada V6T IWS. SUMMARY The role of human chromosome 8 in genetic disease together with the current status of the genetic linkage map for this chromosome is reviewed. Both hereditary genetic disease attributed to mutant alleles at gene loci on chromosome 8 and neoplastic disease owing to somatic mutation, particularly chromosomal translocations, are discussed. Human chromosome 8 is perhaps best known for its In an era when complete sequencing of the human involvement in Burkitt's lymphoma and as the genome is being proposed, it is appropriate for location of the tissue plasminogen activator gene, medical geneticists to accept the challenge of defining by copyright. PLAT, which has been genetically engineered to the set of loci that have mutant alleles causing provide a natural fibrinolytic product for emergency hereditary disease. The fundamental genetic tool of use in cardiac disease. Since chromosome 8 repre- linkage mapping can now be applied, owing largely sents about 5% of the human genome, we may to progress in defining RFLP markers.3 4 This expect it to carry about 5% of human gene loci. This review will focus on genetic disease associated with would correspond to about 90 of the fully validated chromosome 8 loci and the status ofthe chromosome 8 phenotypes in the MIM7 catalogue.' The 27 genes linkage map. -

Naked Mole Rats Can Undergo Developmental, Oncogene-Induced and DNA Damage-Induced Cellular Senescence
Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence Yang Zhaoa,1, Alexander Tyshkovskiyb,c,1, Daniel Muñoz-Espínd,e, Xiao Tiana, Manuel Serranod,f,g, Joao Pedro de Magalhaesh, Eviatar Nevoi,2, Vadim N. Gladyshevc, Andrei Seluanova,2, and Vera Gorbunovaa,2 aDepartment of Biology, University of Rochester, Rochester, NY 14627; bCenter for Data-Intensive Biomedicine and Biotechnology, Skolkovo Institute of Science and Technology, 143028 Moscow, Russia; cDivision of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; dTumor Suppression Group, Spanish National Cancer Research Centre, 28029 Madrid, Spain; eCancer Research UK Cambridge Centre Early Detection Programme, Department of Oncology, University of Cambridge, Hutchison/MRC Research Centre, Cambridge CB2 0XZ, United Kingdom; fInstitute for Research in Biomedicine, Barcelona Institute of Science and Technology, 08028 Barcelona, Spain; gCatalan Institute of Advanced Studies, 08010 Barcelona, Spain; hIntegrative Genomics of Ageing Group, Institute of Ageing and Chronic Disease, University of Liverpool, Liverpool L7 8TX, United Kingdom; and iInstitute of Evolution, University of Haifa, 3498838 Haifa, Israel Contributed by Eviatar Nevo, December 28, 2017 (sent for review December 14, 2017; reviewed by Vadim Fraifeld and Stephen L. Helfand) Cellular senescence is an important anticancer mechanism that contribute to longevity and cancer resistance of the NMR. Our restricts proliferation of damaged or premalignant cells. Cellular earlier studies revealed that cultured NMR fibroblasts exhibit senescence also plays an important role in tissue remodeling early contact inhibition (ECI) (14), which contributes to its during development. However, there is a trade-off associated cancer resistance. ECI could be abrogated by the removal of with cellular senescence as senescent cells contribute to aging high-molecular-mass hyaluronan (15).