The Probability of Parentage Exclusion Based on Restriction Fragment Length Polymorphisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Haploid Genetic Screens Identify an Essential Role for PLP2 in the Downregulation of Novel Plasma Membrane Targets by Viral E3 Ubiquitin Ligases
Haploid Genetic Screens Identify an Essential Role for PLP2 in the Downregulation of Novel Plasma Membrane Targets by Viral E3 Ubiquitin Ligases Richard T. Timms1, Lidia M. Duncan1, Iva A. Tchasovnikarova1, Robin Antrobus1, Duncan L. Smith2, Gordon Dougan3, Michael P. Weekes1, Paul J. Lehner1* 1 Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Cambridge, United Kingdom, 2 Paterson Institute for Cancer Research, University of Manchester, Withington, Manchester, United Kingdom, 3 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, United Kingdom Abstract The Kaposi’s sarcoma-associated herpesvirus gene products K3 and K5 are viral ubiquitin E3 ligases which downregulate MHC-I and additional cell surface immunoreceptors. To identify novel cellular genes required for K5 function we performed a forward genetic screen in near-haploid human KBM7 cells. The screen identified proteolipid protein 2 (PLP2), a MARVEL domain protein of unknown function, as essential for K5 activity. Genetic loss of PLP2 traps the viral ligase in the endoplasmic reticulum, where it is unable to ubiquitinate and degrade its substrates. Subsequent analysis of the plasma membrane proteome of K5-expressing KBM7 cells in the presence and absence of PLP2 revealed a wide range of novel K5 targets, all of which required PLP2 for their K5-mediated downregulation. This work ascribes a critical function to PLP2 for viral ligase activity and underlines the power of non-lethal haploid genetic screens in human cells to identify the genes involved in pathogen manipulation of the host immune system. Citation: Timms RT, Duncan LM, Tchasovnikarova IA, Antrobus R, Smith DL, et al. (2013) Haploid Genetic Screens Identify an Essential Role for PLP2 in the Downregulation of Novel Plasma Membrane Targets by Viral E3 Ubiquitin Ligases. -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

PLP2 (NM 002668) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RG222024 PLP2 (NM_002668) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: PLP2 (NM_002668) Human Tagged ORF Clone Tag: TurboGFP Symbol: PLP2 Synonyms: A4; A4LSB Vector: pCMV6-AC-GFP (PS100010) E. coli Selection: Ampicillin (100 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RG222024 representing NM_002668 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGCGGATTCTGAGCGCCTCTCGGCTCCTGGCTGCTGGGCCGCCTGCACCAACTTCTCGCGCACTCGAA AGGGAATCCTCCTGTTTGCTGAGATTATATTATGCCTGGTGATCCTGATCTGCTTCAGTGCCTCCACACC AGGCTACTCCTCCCTGTCGGTGATTGAGATGATCCTTGCTGCTATTTTCTTTGTTGTCTACATGTGTGAC CTGCACACCAAGATACCATTCATCAACTGGCCCTGGAGTGATTTCTTCCGAACCCTCATAGCGGCAATCC TCTACCTGATCACCTCCATTGTTGTCCTTGTTGAGAGAGGAAACCACTCCAAAATCGTCGCAGGGGTACT GGGCCTAATCGCTACGTGCCTCTTTGGCTATGATGCCTATGTCACCTTCCCCGTTCGGCAGCCAAGACAT ACAGCAGCCCCCACTGACCCCGCAGATGGCCCGGTG ACGCGTACGCGGCCGCTCGAG - GFP Tag - GTTTAA Protein Sequence: >RG222024 representing NM_002668 Red=Cloning site Green=Tags(s) MADSERLSAPGCWAACTNFSRTRKGILLFAEIILCLVILICFSASTPGYSSLSVIEMILAAIFFVVYMCD LHTKIPFINWPWSDFFRTLIAAILYLITSIVVLVERGNHSKIVAGVLGLIATCLFGYDAYVTFPVRQPRH TAAPTDPADGPV TRTRPLE - GFP Tag - V Restriction Sites: SgfI-MluI This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -

Transcriptional Integration of Mitogenic and Mechanical Signals by Myc and YAP
Downloaded from genesdev.cshlp.org on September 30, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH COMMUNICATION rum-mediated and growth factor-mediated cell cycle Transcriptional integration entry (Kelly et al. 1983; Armelin et al. 1984; Roussel of mitogenic and mechanical et al. 1991; Barone and Courtneidge 1995; de Alboran et al. 2001; Trumpp et al. 2001; Perna et al. 2012). This signals by Myc and YAP function of Myc stems from its ability to control the ex- pression of a large fraction of genes involved in cell activa- Ottavio Croci,1,5 Serena De Fazio,1,5 1,5 1,4,5 tion and proliferation. Francesca Biagioni, Elisa Donato, When ectopically expressed in quiescent cells, Myc is Marieta Caganova,1 Laura Curti,1 Mirko Doni,2 able to drive cell cycle progression in the absence of serum Silvia Sberna,1 Deborah Aldeghi,1 (Eilers et al. 1991; Pelengaris et al. 1999). This effect of Chiara Biancotto,1 Alessandro Verrecchia,2 Myc is context-dependent, however, since not all cells or tissues respond to Myc by entering the cell cycle (Jack- Daniela Olivero,3 Bruno Amati,1,2 1 son et al. 1990; Xiao et al. 2001; Murphy et al. 2008). This and Stefano Campaner suggests that a full proliferative response may require the engagement of other TFs, which may respond to different 1Center for Genomic Science of IIT@SEMM (Istituto Italiano di regulatory signals, such as metabolic or mechanical cues. Tecnologia at European School of Molecular Medicine), Recently, YAP has emerged as a key TF in the control of Fondazione Istituto Italiano di Tecnologia (IIT), 20139 Milan, cell growth and organ size in response to a variety of Italy; 2Department of Experimental Oncology, European signals such as cell adhesion, apico–basolateral polarity, Institute of Oncology (IEO), 20139 Milan, Italy; 3Laboratorio di cytoskeletal tension, and mitogens. -

Your Genes, Your Choices
Your Genes, Your Choices: Exploring the Issues Raised by Genetic Research by Catherine Baker Table of Contents Acknowledgments . 6 Introduction . 7 Chapter 1 Martin Needs Medical Treatment (or does he?) . 9 Chapter 2 Priya Should Find Out if She Has Inherited a Fatal Disease (or should she?) . 14 Chapter 3 Howard’s Health Is Up to Him (or is it?) . 26 Chapter 4 Carlos and Mollie Can Have a Perfectly Healthy Baby (or can they?) . 35 Chapter 5 Donita Should Cooperate with the Police (or should she?) . 45 Chapter 6 John and Elsa Will Profit from Biotech Farming (or will they?) . 52 Chapter 7 Dr. Lu’s Patients Have the Right to Be Tall (or do they?) . 62 Chapter 8 Mrs. Fister Can Replace Her Dying Son (or can she?) . 70 Glossary . 81 References . 89 Credits . 81 Science + Literacy for Health Human Genome Project Advisory Board . 93 5 Acknowledgments I am not a science writer by trade. In order to write this book, I first had to study up on genetics and the issues involved. Then I had to try to explain them in a way that other newcomers to the subject could understand, without making terrible errors. It was a difficult task! I am therefore indebted to the members of the AAAS Advisory Panel (listed on page 82). At an all-day meeting in the spring of 1995, they steered my away from my original outline toward the book you find here. Many months later, several panel members provided very useful reviews of the manuscript. For this, I would like to thank Ruth Allen, Jeffrey Botkin, Ron Cole-Turner, Robert Cook-Deegan, and Joan Weiss. -

Structure and Mechanism of the RNA Polymerase II Transcription Machinery
Downloaded from genesdev.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Structure and mechanism of the RNA polymerase II transcription machinery Allison C. Schier and Dylan J. Taatjes Department of Biochemistry, University of Colorado, Boulder, Colorado 80303, USA RNA polymerase II (Pol II) transcribes all protein-coding ingly high resolution, which has rapidly advanced under- genes and many noncoding RNAs in eukaryotic genomes. standing of the molecular basis of Pol II transcription. Although Pol II is a complex, 12-subunit enzyme, it lacks Structural biology continues to transform our under- the ability to initiate transcription and cannot consistent- standing of complex biological processes because it allows ly transcribe through long DNA sequences. To execute visualization of proteins and protein complexes at or near these essential functions, an array of proteins and protein atomic-level resolution. Combined with mutagenesis and complexes interact with Pol II to regulate its activity. In functional assays, structural data can at once establish this review, we detail the structure and mechanism of how enzymes function, justify genetic links to human dis- over a dozen factors that govern Pol II initiation (e.g., ease, and drive drug discovery. In the past few decades, TFIID, TFIIH, and Mediator), pausing, and elongation workhorse techniques such as NMR and X-ray crystallog- (e.g., DSIF, NELF, PAF, and P-TEFb). The structural basis raphy have been complemented by cryoEM, cross-linking for Pol II transcription regulation has advanced rapidly mass spectrometry (CXMS), and other methods. Recent in the past decade, largely due to technological innova- improvements in data collection and imaging technolo- tions in cryoelectron microscopy. -

The Ubiquitin Proteasome System Is Required for Cell Proliferation of The
RESEARCH ARTICLE 3257 Development 137, 3257-3268 (2010) doi:10.1242/dev.053124 © 2010. Published by The Company of Biologists Ltd The ubiquitin proteasome system is required for cell proliferation of the lens epithelium and for differentiation of lens fiber cells in zebrafish Fumiyasu Imai1, Asuka Yoshizawa1, Noriko Fujimori-Tonou2, Koichi Kawakami3 and Ichiro Masai1,* SUMMARY In the developing vertebrate lens, epithelial cells differentiate into fiber cells, which are elongated and flat in shape and form a multilayered lens fiber core. In this study, we identified the zebrafish volvox (vov) mutant, which shows defects in lens fiber differentiation. In the vov mutant, lens epithelial cells fail to proliferate properly. Furthermore, differentiating lens fiber cells do not fully elongate, and the shape and position of lens fiber nuclei are affected. We found that the vov mutant gene encodes Psmd6, the subunit of the 26S proteasome. The proteasome regulates diverse cellular functions by degrading polyubiquitylated proteins. Polyubiquitylated proteins accumulate in the vov mutant. Furthermore, polyubiquitylation is active in nuclei of differentiating lens fiber cells, suggesting roles of the proteasome in lens fiber differentiation. We found that an E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) is involved in lens defects in the vov mutant. These data suggest that the ubiquitin proteasome system is required for cell proliferation of lens epithelium and for the differentiation of lens fiber cells in zebrafish. KEY WORDS: Ubiquitin, Proteasome, Lens, Zebrafish, Psmd6, APC/C INTRODUCTION acid DNase (DLAD; Dnase2b). However, it is unclear how lens During development, the lens placode delaminates from the morphogenesis and organelle loss are related to these transcription epidermal ectoderm and forms the lens vesicle. -

Proteolipoprotein Gene Analysis in 82 Patients with Sporadic Pelizaeus
Am. J. Hum. Genet. 65:360±369, 1999 Proteolipoprotein Gene Analysis in 82 Patients with Sporadic Pelizaeus-Merzbacher Disease: Duplications, the Major Cause of the Disease, Originate More Frequently in Male Germ Cells, but Point Mutations Do Not Corinne Mimault,1 GenevieÁve Giraud,1 Virginie Courtois,1 Fabrice Cailloux,1 Jean Yves Boire,2 Bernard Dastugue,1 Odile Boesp¯ug-Tanguy,1 and the Clinical European Network on Brain Dysmyelinating Disease* 1INSERM U.384, FaculteÂdeMeÂdecine and 2ERIM, FaculteÂdeMeÂdecine, Clermont-Ferrand, France Summary Introduction Pelizaeus-Merzbacher Disease (PMD) is an X-linked de- Pelizaeus-Merzbacher disease (PMD; MIM 312080), is velopmental defect of myelination affecting the central an X-linked defect of myelin formation affecting the nervous system and segregating with the proteolipopro- CNS. Originally described by Pelizaeus (1885), the clin- tein (PLP) locus. Investigating 82 strictly selected spo- ical syndrome was neuropathologically de®ned by Merz- radic cases of PMD, we found PLP mutations in 77%; bacher (1910) as a diffuse hypomyelination of the CNS, complete PLP-gene duplications were the most frequent associated with an abnormally low number of mature abnormality (62%), whereas point mutations in coding oligodendrocytes. The diagnosis is based on early/im- or splice-site regions of the gene were involved less fre- paired motor development (during the ®rst 3 mo of life) quently (38%). We analyzed the maternal status of 56 characterized by severe hypotonia associated with nys- cases to determine the origin of both types of PLP mu- tagmus and, later, the development of abnormal move- tation, since this is relevant to genetic counseling. -

"The Genecards Suite: from Gene Data Mining to Disease Genome Sequence Analyses". In: Current Protocols in Bioinformat
The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses Gil Stelzer,1,5 Naomi Rosen,1,5 Inbar Plaschkes,1,2 Shahar Zimmerman,1 Michal Twik,1 Simon Fishilevich,1 Tsippi Iny Stein,1 Ron Nudel,1 Iris Lieder,2 Yaron Mazor,2 Sergey Kaplan,2 Dvir Dahary,2,4 David Warshawsky,3 Yaron Guan-Golan,3 Asher Kohn,3 Noa Rappaport,1 Marilyn Safran,1 and Doron Lancet1,6 1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel 2LifeMap Sciences Ltd., Tel Aviv, Israel 3LifeMap Sciences Inc., Marshfield, Massachusetts 4Toldot Genetics Ltd., Hod Hasharon, Israel 5These authors contributed equally to the paper 6Corresponding author GeneCards, the human gene compendium, enables researchers to effectively navigate and inter-relate the wide universe of human genes, diseases, variants, proteins, cells, and biological pathways. Our recently launched Version 4 has a revamped infrastructure facilitating faster data updates, better-targeted data queries, and friendlier user experience. It also provides a stronger foundation for the GeneCards suite of companion databases and analysis tools. Improved data unification includes gene-disease links via MalaCards and merged biological pathways via PathCards, as well as drug information and proteome expression. VarElect, another suite member, is a phenotype prioritizer for next-generation sequencing, leveraging the GeneCards and MalaCards knowledgebase. It au- tomatically infers direct and indirect scored associations between hundreds or even thousands of variant-containing genes and disease phenotype terms. Var- Elect’s capabilities, either independently or within TGex, our comprehensive variant analysis pipeline, help prepare for the challenge of clinical projects that involve thousands of exome/genome NGS analyses. -

The General Transcription Factors of RNA Polymerase II
Downloaded from genesdev.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW The general transcription factors of RNA polymerase II George Orphanides, Thierry Lagrange, and Danny Reinberg 1 Howard Hughes Medical Institute, Department of Biochemistry, Division of Nucleic Acid Enzymology, Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey, Piscataway, New Jersey 08854-5635 USA Messenger RNA (mRNA) synthesis occurs in distinct unique functions and the observation that they can as- mechanistic phases, beginning with the binding of a semble at a promoter in a specific order in vitro sug- DNA-dependent RNA polymerase to the promoter re- gested that a preinitiation complex must be built in a gion of a gene and culminating in the formation of an stepwise fashion, with the binding of each factor promot- RNA transcript. The initiation of mRNA transcription is ing association of the next. The concept of ordered as- a key stage in the regulation of gene expression. In eu- sembly recently has been challenged, however, with the karyotes, genes encoding mRNAs and certain small nu- discovery that a subset of the GTFs exists in a large com- clear RNAs are transcribed by RNA polymerase II (pol II). plex with pol II and other novel transcription factors. However, early attempts to reproduce mRNA transcrip- The existence of this pol II holoenzyme suggests an al- tion in vitro established that purified pol II alone was not ternative to the paradigm of sequential GTF assembly capable of specific initiation (Roeder 1976; Weil et al. (for review, see Koleske and Young 1995). -
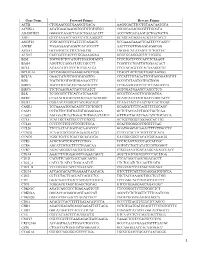
Gene Name Forward Primer Reverse Primer ACTB
Gene Name Forward Primer Reverse Primer ACTB CTGGAACGGTGAAGGTGACA AAGGGACTTCCTGTAACAATGCA ACVRL1 ACATGAAGAAGGTGGTGTGTGTGG CGGGCAGAGGGGTTTGGGTA ADAMDEC1 GGGGCCAGACTACACTGAAACATT ACCCGTCACAAGTACTGATGCTG AHI1 GTCCAAAACTACCCCATCAAGGCT GCAGCACAGGAACGTATCACCT ANGPT2 TGGCAGCGTTGATTTTCAGAGG GCGAAACAAACTCATTTCCCAGCC ANPEP TGAAGAAGCAGGTCACACCCCT AACTCCGTTGGAGCAGGCGG APOA1 GCCGTGCTCTTCCTGACGG TGGGACACATAGTCTCTGCCGC ATXN7 CACCGCCCACTCTGGAAAAGAA GGGTGCAGGGCTTCTTGGTG B2M TGCTGTCTCCATGTTTGATGTATCT TCTCTGCTCCCCACCTCTAAGT BAG4 AGGTTCCAGGATATCCGCCTT TCGGTCCTGATTGTGGAACACT BCL2 ACAACATCGCCCTGTGGATGA CCGTACAGTTCCACAAAGGCAT BCL2L14 GCTCAGGGTCAAAGGACGTTGG TCAGCTACTCGGTTGGCAATGG BCL7A GAACCATGTCGGGCAGGTCG CCCATTTGTAGATTCGTAGGGATGTGT BIN1 TGCTGTCGTGGTGGAGACCTTC GCCGTGTAGTCGTGCTGGG BIRC3 TGCTATCCACATCAGACAGCCC TCTGAATGGTCTTCTCCAGGTTCA BIRC5 TTCTCAAGGACCACCGCATCT AGTGGATGAAGCCAGCCTCG BLK TCGGGGTCTTCACCATCAAAGC GCGCTCCAGGTTGCGGATGA BTRC CCAAATGTGTCATTACCAACATGGGC GCAGCACATAGTGATTTGGCATCC BUB3 CGGAACATGGGTTACGTGCAGC CCAAATACTCAACTGCCACTCGGC CAGE1 TCCAAAATGCACAGTCTTCTGGCT GGAGGCTCTTCAGTTTTTGCAGC CASP1 CCTGTTCCTGTGATGTGGAGGAAA GCTCTACCATCTGGCTGCTCAA CASP3 AGCGAATCAATGGACTCTGGAATATCC GTTTGCTGCATCGACATCTGTACCA CCL5 TCATTGCTACTGCCCTCTGCG ACTGCTGGGTTGGAGCACTTG CCL18 CCCTCCTTGTCCTCGTCTGCA GCACTGGGGGCTGGTTTCAG CCL26 TTCCAATACAGCCACAAGCCCC GGATGGGTACAGACTTTCTTGCCTC CCND2 TCAAGTGCGTGCAGAAGGACAT CTTCGCACTTCTGTTCCTCACA CCND3 TGGCTGCTGTGATTGCACATGA GATGGCGGGTACATGGCAAAGG CCR3 ACGCTGCTCTGCTTCCTGG TCCTCAGTTCCCCACCATCGC CCR4 AGCATCGTGCTTCCTGAGCAA GGTGTCTGCTATATCCGTGGGGT CCR7 AGACAGGGGTAGTGCGAGGC