Multiple QTL Underlie Milk Phenotypes at the CSF2RB Locus Thomas J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Multistage Analysis of Variants in the Inflammation Pathway and Lung Cancer Risk in Smokers
Published OnlineFirst May 9, 2012; DOI: 10.1158/1055-9965.EPI-12-0352-T Cancer Epidemiology, Research Article Biomarkers & Prevention Multistage Analysis of Variants in the Inflammation Pathway and Lung Cancer Risk in Smokers Margaret R. Spitz1, Ivan P. Gorlov2, Qiong Dong3, Xifeng Wu3, Wei Chen4, David W. Chang3, Carol J. Etzel3, Neil E. Caporaso5, Yang Zhao8, David C. Christiani8, Paul Brennan9, Demetrius Albanes7, Jianxin Shi6, Michael Thun10, Maria Teresa Landi5, and Christopher I. Amos4 Abstract Background: Tobacco-induced lung cancer is characterized by a deregulated inflammatory microenviron- ment. Variants in multiple genes in inflammation pathways may contribute to risk of lung cancer. Methods: We therefore conducted a three-stage comprehensive pathway analysis (discovery, replication, and meta-analysis) of inflammation gene variants in ever-smoking lung cancer cases and controls. A discovery set (1,096 cases and 727 controls) and an independent and nonoverlapping internal replication set (1,154 cases and 1,137 controls) were derived from an ongoing case–control study. For discovery, we used an iSelect BeadChip to interrogate a comprehensive panel of 11,737 inflammation pathway single-nucleotide poly- morphisms (SNP) and selected nominally significant (P < 0.05) SNPs for internal replication. Results: There were six SNPs that achieved statistical significance (P < 0.05) in the internal replication data set with concordant risk estimates for former smokers and five concordant and replicated SNPs in current smokers. Replicated hits were further tested in a subsequent meta-analysis using external data derived from two published genome-wide association studies (GWAS) and a case–control study. Two of these variants (a BCL2L14 SNP in former smokers and an SNP in IL2RB in current smokers) were further validated. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Contribution of IL9, IL2RA and IL2RB Genetic Polymorphisms in Coronary Heart Disease in Chinese Han Population
Contribution of IL9, IL2RA and IL2RB genetic polymorphisms in coronary heart disease in Chinese Han population Xianghong Chen The Second Aliated Hospital of Hainan Medical University Xingfan Wang The Second Aliated Hospital of Hainan Medical University Zaozhang q Zhang The second Aliated Hospital of Hainan Medical University Yuewu Chen The Second Aliated Hospital of Hainan Medical University Chao Wang ( [email protected] ) The Second Aliated Hospital of Hainan Medical Universiy https://orcid.org/0000-0001-5632-9778 Research article Keywords: Posted Date: December 9th, 2019 DOI: https://doi.org/10.21203/rs.2.18401/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/11 Abstract Background: Coronary heart disease (CHD) is one of the leading causes of disability and death worldwide. In the pathogenesis of CHD, inammatory cytokines take an essential part. This study was designed to detect the potential association between IL-9, IL-2RA and IL-2RB variants and CHD in Chinese Han population. Methods: This case-control study conducted 499 CHD patients and 496 healthy controls. Seven selected SNPs were genotyped to investigate the possible association between the polymorphisms and the CHD risk. The interaction of SNP-SNP in the CHD risk was analyzed by Multifactor dimensionality reduction (MDR). Results: We observed an association between IL-9 rs55692658 (OR = 1.72, p = 0.003) and the increased CHD risk. The stratication analysis by age indicated that no matter participants who were older or younger than 61 years, IL-9 rs55692658 and IL-2RB rs1573673 contributed to the CHD susceptibility signicantly (p < 0.05, respectively). -
GM-CSF Receptor-Beta, Human, Recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor Beta
GM-CSF Receptor-beta, human, recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor beta Instruction Manual Catalog Number C-60430 Synonyms CSF2RB,Colony Stimulating Factor 2 Receptor, Beta, Low-Affinity (Granulocyte-Macrophage), GM-CSF/IL-3/IL-5 Receptor Common Beta Subunit, CDw131, IL3RB, SMDP5, IL5RB, Interleukin 3 Receptor/Granulocyte-Macrophage Colony Stimulating Factor 3 Receptor, Beta (High Affinity), Colony-Stimulating Factor-2 Receptor, Beta, Low-Affinity, GM-CSF/IL-3/IL-5 Receptor Common Beta-Chain, Cytokine Receptor Common Subunit Beta, CD131 Antigen, CD131 Description GM-CSF Receptor-beta, also known as CSF2RB is a member of the type I cytokine receptor family. CSF2RB is a high affinity receptor for interleukin-3, interleukin-5 as well as granulocyte- macrophage colony-stimulating factor. The CSF2RB unique form of receptor assembly applies also to IL-3 and IL-5 receptors, providing a structural basis for understanding their activation mechanism which is essential for the development of therapeutics. Human recombinant GM-CSF Receptor-beta produced in Sf9 cells is a single, glycosylated polypeptide chain of 435 amino acids (17-443 a.a), having a molecular mass of 49.7 kDa (migrates at 40-57 kDa on SDS-PAGE under reducing conditions). CSF2RB is fused to an 8 amino acid His-tag at the C-terminus and purified using proprietary chromatographic techniques. Quantity 10 µg Molecular Mass 49.7 kDa Source Sf9 insect cells Biological-Activity NA Specific Activity NA Formulation Sterile-filtered, clear protein solution (0.5 mg/ml) containing phosphate-buffered saline (pH 7.4) and 10% glycerol. Reconstitution Please Note: Always centrifuge product briefly before opening vial. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -
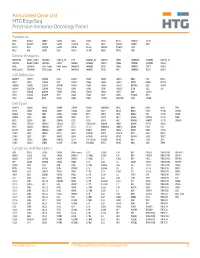
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2 -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

Conserved Transcriptomic Profile Between Mouse and Human Colitis
ARTICLE https://doi.org/10.1038/s41467-019-10769-x OPEN Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification Paulo Czarnewski1, Sara M. Parigi1, Chiara Sorini 1, Oscar E. Diaz 1, Srustidhar Das1, Nicola Gagliani1,2,3 & Eduardo J. Villablanca 1,3 1234567890():,; Clinical manifestations and response to therapies in ulcerative colitis (UC) are hetero- geneous, yet patient classification criteria for tailored therapies are currently lacking. Here, we present an unsupervised molecular classification of UC patients, concordant with response to therapy in independent retrospective cohorts. We show that classical clustering of UC patient tissue transcriptomic data sets does not identify clinically relevant profiles, likely due to associated covariates. To overcome this, we compare cross-sectional human data sets with a newly generated longitudinal transcriptome profile of murine DSS-induced colitis. We show that the majority of colitis risk-associated gene expression peaks during the inflammatory rather than the recovery phase. Moreover, we achieve UC patient clustering into two distinct transcriptomic profiles, differing in neutrophil-related gene activation. Notably, 87% of patients in UC1 cluster are unresponsive to two most widely used biological therapies. These results demonstrate that cross-species comparison enables stratification of patients undistinguishable by other molecular approaches. 1 Immunology and Allergy Unit, Department of Medicine, Solna, Karolinska Institute and University Hospital, 17176 Stockholm, Sweden. 2 Department of Medicine and Department of General, Visceral and Thoracic Surgery, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany. 3These authors jointly supervised this work: Nicola Gagliani, Eduardo J. Villablanca. Correspondence and requests for materials should be addressed to E.J.V. -

Tethering IL2 to Its Receptor Il2rb Enhances Antitumor Activity and Expansion of Natural Killer NK92 Cells Youssef Jounaidi, Joseph F
Published OnlineFirst September 15, 2017; DOI: 10.1158/0008-5472.CAN-17-1007 Cancer Therapeutics, Targets, and Chemical Biology Research Tethering IL2 to Its Receptor IL2Rb Enhances Antitumor Activity and Expansion of Natural Killer NK92 Cells Youssef Jounaidi, Joseph F. Cotten, Keith W. Miller, and Stuart A. Forman Abstract IL2 is an immunostimulatory cytokine for key immune cells of IL2 and its receptor IL2Rb joined via a peptide linker (CIRB). including T cells and natural killer (NK) cells. Systemic IL2 NK92 cells expressing CIRB (NK92CIRB) were highly activated and supplementation could enhance NK-mediated immunity in a expanded indefinitely without exogenous IL2. When compared variety of diseases ranging from neoplasms to viral infection. with an IL2-secreting NK92 cell line, NK92CIRB were more acti- However, its systemic use is restricted by its serious side effects and vated, cytotoxic, and resistant to growth inhibition. Direct contact limited efficacy due to activation of T regulatory cells (Tregs). IL2 with cancer cells enhanced the cytotoxic character of NK92CIRB signaling is mediated through interactions with a multi-subunit cells, which displayed superior in vivo antitumor effects in mice. receptor complex containing IL2Ra, IL2Rb, and IL2Rg. Adult Overall, our results showed how tethering IL2 to its receptor natural killer (NK) cells express only IL2Rb and IL2Rg subunits IL2Rb eliminates the need for IL2Ra and IL2Rb, offering a and are therefore relatively insensitive to IL2. To overcome these new tool to selectively activate and empower immune therapy. limitations, we created a novel chimeric IL2-IL2Rb fusion protein Cancer Res; 77(21); 5938–51. Ó2017 AACR. Introduction in order for allogeneic NK cells to be effective, pretransfer lymphodepletion is required to reduce competition for growth Natural killer (NK) cells are lymphocytes endowed with the factors and cytokines (14, 15). -

A Low Interleukin-2 Receptor Signaling Threshold Supports the Development and Homeostasis of T Regulatory Cells
Immunity Article A Low Interleukin-2 Receptor Signaling Threshold Supports the Development and Homeostasis of T Regulatory Cells Aixin Yu,1 Linjian Zhu,1 Norman H. Altman,2 and Thomas R. Malek1,3,* 1Department of Microbiology and Immunology 2Department of Pathology 3Diabetes Research Institute Miller School of Medicine, University of Miami, P.O. Box 01960, Miami, FL 33101, USA *Correspondence: [email protected] DOI 10.1016/j.immuni.2008.11.014 SUMMARY of the genetic factors rendering nonobese diabetic (NOD) mice susceptible to autoimmune disease that leads to impaired Treg Interleukin-2 receptor (IL-2R) signaling is essential cells (Yamanouchi et al., 2007). IL-2 is also necessary for the for T regulatory (Treg) cell development and homeo- generation of Foxp3+ induced-Treg (iTreg) cells from naive stasis. Here, we show that expression of IL-2Rb conventional peripheral T lymphocytes (Davidson et al., 2007; chains that lack tyrosine residues important for the Zheng et al., 2007). association of the adaptor Shc and the transcription IL-2R signaling mechanisms have been most extensively factor STAT5 in IL-2Rb-deficient mice resulted in studied in various IL-2-responsive cell lines that approximate activated effector T cells (Gaffen, 2001; Nelson and Willerford, production of a normal proportion of natural Treg 1998). IL-2 signal transduction is initiated upon ligand-induced cells that suppressed severe autoimmunity related oligomerization of the IL-2R, consisting of IL-2Ra, IL-2Rb, and with deficiency in IL-2 or IL-2R. These mutant IL- gc (CD132). This event brings the cytoplasmic tail of IL-2Rb and 2Rb chains supported suboptimal and transient gc in close proximity with their associated Jak-1 and Jak-3 STAT5 activation that upregulate the transcription kinases, respectively, allowing phosphorylation of three key factor Foxp3 to normal amounts in natural, but not tyrosines (Y) residues of IL-2Rb. -

Calcium-Mediated Shaping of Naive CD4 T-Cell Phenotype and Function
RESEARCH ARTICLE Calcium-mediated shaping of naive CD4 T-cell phenotype and function Vincent Guichard1,2, Nelly Bonilla1, Aure´ lie Durand1, Alexandra Audemard-Verger1, Thomas Guilbert1, Bruno Martin1, Bruno Lucas1†, Ce´ dric Auffray1†* 1Institut Cochin, Paris Descartes Universite´, CNRS UMR8104, INSERM U1016, Paris, France; 2Paris Diderot Universite´, Paris, France Abstract Continuous contact with self-major histocompatibility complex ligands is essential for the survival of naive CD4 T cells. We have previously shown that the resulting tonic TCR signaling also influences their fate upon activation by increasing their ability to differentiate into induced/ peripheral regulatory T cells. To decipher the molecular mechanisms governing this process, we here focus on the TCR signaling cascade and demonstrate that a rise in intracellular calcium levels is sufficient to modulate the phenotype of mouse naive CD4 T cells and to increase their sensitivity to regulatory T-cell polarization signals, both processes relying on calcineurin activation. Accordingly, in vivo calcineurin inhibition leads the most self-reactive naive CD4 T cells to adopt the phenotype of their less self-reactive cell-counterparts. Collectively, our findings demonstrate that calcium- mediated activation of the calcineurin pathway acts as a rheostat to shape both the phenotype and effector potential of naive CD4 T cells in the steady-state. DOI: https://doi.org/10.7554/eLife.27215.001 *For correspondence: Introduction [email protected] T-cell precursors originate in the bone-marrow and are educated in the thymus through processes † called positive and negative selections, which result in MHC-restriction and self-tolerance, respec- These authors contributed equally to this work tively (Stritesky et al., 2012). -

Molecular Biology of Hodgkin Lymphoma
Leukemia (2021) 35:968–981 https://doi.org/10.1038/s41375-021-01204-6 REVIEW ARTICLE Lymphoma Molecular biology of Hodgkin lymphoma 1 1 Marc A. Weniger ● Ralf Küppers Received: 17 November 2020 / Revised: 1 February 2021 / Accepted: 18 February 2021 / Published online: 8 March 2021 © The Author(s) 2021. This article is published with open access Abstract Classical Hodgkin lymphoma (cHL) is unique among lymphoid malignancies in several key biological features. (i) The Hodgkin and Reed-Sternberg (HRS) tumor cells are rare among an extensive and complex microenvironment. (ii) They derive from B cells, but have largely lost the B-cell typical gene expression program. (iii) Their specific origin appears to be pre-apoptotic germinal center (GC) B cells. (iv) They consistently develop bi- or multinucleated Reed-Sternberg cells from mononuclear Hodgkin cells. (v) They show constitutive activation of numerous signaling pathways. Recent studies have begun to uncover the basis of these specific features of cHL: HRS cells actively orchestrate their complex microenvironment and attract many distinct subsets of immune cells into the affected tissues, to support their survival and proliferation, and to create an immunosuppressive environment. Reed-Sternberg cells are generated by incomplete cytokinesis and refusion of Hodgkin cells. Epstein-Barr virus (EBV) plays a major role in the rescue of crippled GC B cells 1234567890();,: 1234567890();,: from apoptosis and hence is a main player in early steps of lymphomagenesis of EBV+ cHL cases. The analysis of the landscape of genetic lesions in HRS cells so far did not reveal any highly recurrent HRS cell-specific lesions, but major roles of genetic lesions in members of the NF-κB and JAK/STAT pathways and of factors of immune evasion.