Foxp3-Independent Mechanism by Which TGF-Β Controls Peripheral T Cell Tolerance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
(12) Patent Application Publication (10) Pub. No.: US 2012/0070450 A1 Ishikawa Et Al
US 20120070450A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0070450 A1 Ishikawa et al. (43) Pub. Date: Mar. 22, 2012 (54) LEUKEMA STEM CELLMARKERS Publication Classification (51) Int. Cl. A 6LX 39/395 (2006.01) (75) Inventors: Fumihiko Ishikawa, Kanagawa CI2O I/68 (2006.01) (JP): Osamu Ohara, Kanagawa GOIN 2L/64 (2006.01) (JP); Yoriko Saito, Kanagawa (JP); A6IP35/02 (2006.01) Hiroshi Kitamura, Kanagawa (JP); C40B 30/04 (2006.01) Atsushi Hijikata, Kanagawa (JP); A63L/7088 (2006.01) Hidetoshi Ozawa, Kanagawa (JP); C07K 6/8 (2006.01) Leonard D. Shultz, Bar Harbor, C7H 2L/00 (2006.01) A6II 35/12 (2006.01) ME (US) CI2N 5/078 (2010.01) (52) U.S. Cl. .................. 424/173.1; 424/178.1; 424/93.7: (73) Assignee: RIKEN, Wako-shi (JP) 435/6.14; 435/723; 435/375; 506/9: 514/44 A: 530/389.6; 530/391.7:536/24.5 (57) ABSTRACT (21) Appl. No.: 13/258,993 The invention provides a test method for predicting the initial onset or a recurrence of acute myeloid leukemia (AML) com PCT Fled: prising (1) measuring the expression level of human leukemic (22) Mar. 24, 2010 stem cell (LSC) marker genes in a biological sample collected from a Subject for a transcription product or translation prod uct of the gene as an analyte and (2) comparing the expression (86) PCT NO.: PCT/UP2010/0551.31 level with a reference value; an LSC-targeting therapeutic agent for AML capable of Suppressing the expression of a S371 (c)(1), gene selected from among LSC marker genes or a Substance (2), (4) Date: Dec. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Role of IL-4 Receptor &Alpha
Role of IL-4 receptor a–positive CD41 T cells in chronic airway hyperresponsiveness Frank Kirstein, PhD, Natalie E. Nieuwenhuizen, PhD,* Jaisubash Jayakumar, PhD, William G. C. Horsnell, PhD, and Frank Brombacher, PhD Cape Town, South Africa, and Berlin, Germany Background: TH2 cells and their cytokines are associated with IL-17–producing T cells and, consequently, increased airway allergic asthma in human subjects and with mouse models of neutrophilia. allergic airway disease. IL-4 signaling through the IL-4 receptor Conclusion: IL-4–responsive T helper cells are dispensable for 1 a (IL-4Ra) chain on CD4 T cells leads to TH2 cell acute OVA-induced airway disease but crucial in maintaining differentiation in vitro, implying that IL-4Ra–responsive CD41 chronic asthmatic pathology. (J Allergy Clin Immunol T cells are critical for the induction of allergic asthma. However, 2016;137:1852-62.) mechanisms regulating acute and chronic allergen-specific T 2 H Key words: responses in vivo remain incompletely understood. TH2 cell, acute allergic airway disease, chronic asthma, Objective: This study defines the requirements for IL-4Ra– cytokine receptors, IL-4, IL-13, gene-deficient mice responsive CD41 T cells and the IL-4Ra ligands IL-4 and IL-13 in the development of allergen-specific TH2 responses during the Allergic asthma is a chronic inflammatory disease of the airways onset and chronic phase of experimental allergic airway disease. characterized by an inappropriate immune response to harmless Methods: Development of acute and chronic ovalbumin environmental antigens. T 2 cells regulate adaptive immune 1 H (OVA)–induced allergic asthma was assessed weekly in CD4 T responses to allergens, and their presence correlates with disease 2 cell–specific IL-4Ra–deficient BALB/c mice (LckcreIL-4Ra /lox) symptoms in human subjects and mice.1 IL-4 plays a crucial role and respective control mice in the presence or absence of IL-4 in the in vitro and in vivo differentiation of TH2 cells, suggesting or IL-13. -

List of Genes Used in Cell Type Enrichment Analysis
List of genes used in cell type enrichment analysis Metagene Cell type Immunity ADAM28 Activated B cell Adaptive CD180 Activated B cell Adaptive CD79B Activated B cell Adaptive BLK Activated B cell Adaptive CD19 Activated B cell Adaptive MS4A1 Activated B cell Adaptive TNFRSF17 Activated B cell Adaptive IGHM Activated B cell Adaptive GNG7 Activated B cell Adaptive MICAL3 Activated B cell Adaptive SPIB Activated B cell Adaptive HLA-DOB Activated B cell Adaptive IGKC Activated B cell Adaptive PNOC Activated B cell Adaptive FCRL2 Activated B cell Adaptive BACH2 Activated B cell Adaptive CR2 Activated B cell Adaptive TCL1A Activated B cell Adaptive AKNA Activated B cell Adaptive ARHGAP25 Activated B cell Adaptive CCL21 Activated B cell Adaptive CD27 Activated B cell Adaptive CD38 Activated B cell Adaptive CLEC17A Activated B cell Adaptive CLEC9A Activated B cell Adaptive CLECL1 Activated B cell Adaptive AIM2 Activated CD4 T cell Adaptive BIRC3 Activated CD4 T cell Adaptive BRIP1 Activated CD4 T cell Adaptive CCL20 Activated CD4 T cell Adaptive CCL4 Activated CD4 T cell Adaptive CCL5 Activated CD4 T cell Adaptive CCNB1 Activated CD4 T cell Adaptive CCR7 Activated CD4 T cell Adaptive DUSP2 Activated CD4 T cell Adaptive ESCO2 Activated CD4 T cell Adaptive ETS1 Activated CD4 T cell Adaptive EXO1 Activated CD4 T cell Adaptive EXOC6 Activated CD4 T cell Adaptive IARS Activated CD4 T cell Adaptive ITK Activated CD4 T cell Adaptive KIF11 Activated CD4 T cell Adaptive KNTC1 Activated CD4 T cell Adaptive NUF2 Activated CD4 T cell Adaptive PRC1 Activated -

Contribution of IL9, IL2RA and IL2RB Genetic Polymorphisms in Coronary Heart Disease in Chinese Han Population
Contribution of IL9, IL2RA and IL2RB genetic polymorphisms in coronary heart disease in Chinese Han population Xianghong Chen The Second Aliated Hospital of Hainan Medical University Xingfan Wang The Second Aliated Hospital of Hainan Medical University Zaozhang q Zhang The second Aliated Hospital of Hainan Medical University Yuewu Chen The Second Aliated Hospital of Hainan Medical University Chao Wang ( [email protected] ) The Second Aliated Hospital of Hainan Medical Universiy https://orcid.org/0000-0001-5632-9778 Research article Keywords: Posted Date: December 9th, 2019 DOI: https://doi.org/10.21203/rs.2.18401/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/11 Abstract Background: Coronary heart disease (CHD) is one of the leading causes of disability and death worldwide. In the pathogenesis of CHD, inammatory cytokines take an essential part. This study was designed to detect the potential association between IL-9, IL-2RA and IL-2RB variants and CHD in Chinese Han population. Methods: This case-control study conducted 499 CHD patients and 496 healthy controls. Seven selected SNPs were genotyped to investigate the possible association between the polymorphisms and the CHD risk. The interaction of SNP-SNP in the CHD risk was analyzed by Multifactor dimensionality reduction (MDR). Results: We observed an association between IL-9 rs55692658 (OR = 1.72, p = 0.003) and the increased CHD risk. The stratication analysis by age indicated that no matter participants who were older or younger than 61 years, IL-9 rs55692658 and IL-2RB rs1573673 contributed to the CHD susceptibility signicantly (p < 0.05, respectively). -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

An Ontogenetic Switch Drives the Positive and Negative Selection of B Cells
An ontogenetic switch drives the positive and negative selection of B cells Xijin Xua, Mukta Deobagkar-Lelea, Katherine R. Bulla, Tanya L. Crockforda, Adam J. Meadb, Adam P. Cribbsc, David Simsc, Consuelo Anzilottia, and Richard J. Cornalla,1 aMedical Research Council Human Immunology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, OX3 9DS Oxford, United Kingdom; bMedical Research Council Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, OX3 9DS Oxford, United Kingdom; and cMedical Research Council, Weatherall Institute of Molecular Medicine, Centre for Computational Biology, Weatherall Institute of Molecular Medicine, University of Oxford, OX3 9DS Oxford, United Kingdom Edited by Michael Reth, University of Freiburg, Freiburg, Germany, and approved January 6, 2020 (received for review September 3, 2019) + Developing B cells can be positively or negatively selected by self- BM HSCs increased CD5 B-1a B cell development (15), while antigens, but the mechanisms that determine these outcomes are expression of let-7b in FL pro-B cells blocked the development of incompletely understood. Here, we show that a B cell intrinsic B-1 B cells (17). These findings support the notion of hard-wired switch between positive and negative selection during ontogeny differences during ontogeny, but possibly downstream of the HSC is determined by a change from Lin28b to let-7 gene expression. commitment stage. Ectopic expression of a Lin28b transgene in murine B cells restored Several lines of evidence also suggest that B-1 B cells can un- the positive selection of autoreactive B-1 B cells by self-antigen in dergo positive selection, which is linked to their B cell receptor adult bone marrow. -

B Cell Checkpoints in Autoimmune Rheumatic Diseases
REVIEWS B cell checkpoints in autoimmune rheumatic diseases Samuel J. S. Rubin1,2,3, Michelle S. Bloom1,2,3 and William H. Robinson1,2,3* Abstract | B cells have important functions in the pathogenesis of autoimmune diseases, including autoimmune rheumatic diseases. In addition to producing autoantibodies, B cells contribute to autoimmunity by serving as professional antigen- presenting cells (APCs), producing cytokines, and through additional mechanisms. B cell activation and effector functions are regulated by immune checkpoints, including both activating and inhibitory checkpoint receptors that contribute to the regulation of B cell tolerance, activation, antigen presentation, T cell help, class switching, antibody production and cytokine production. The various activating checkpoint receptors include B cell activating receptors that engage with cognate receptors on T cells or other cells, as well as Toll-like receptors that can provide dual stimulation to B cells via co- engagement with the B cell receptor. Furthermore, various inhibitory checkpoint receptors, including B cell inhibitory receptors, have important functions in regulating B cell development, activation and effector functions. Therapeutically targeting B cell checkpoints represents a promising strategy for the treatment of a variety of autoimmune rheumatic diseases. Antibody- dependent B cells are multifunctional lymphocytes that contribute that serve as precursors to and thereby give rise to acti- cell- mediated cytotoxicity to the pathogenesis of autoimmune diseases -
GM-CSF Receptor-Beta, Human, Recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor Beta
GM-CSF Receptor-beta, human, recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor beta Instruction Manual Catalog Number C-60430 Synonyms CSF2RB,Colony Stimulating Factor 2 Receptor, Beta, Low-Affinity (Granulocyte-Macrophage), GM-CSF/IL-3/IL-5 Receptor Common Beta Subunit, CDw131, IL3RB, SMDP5, IL5RB, Interleukin 3 Receptor/Granulocyte-Macrophage Colony Stimulating Factor 3 Receptor, Beta (High Affinity), Colony-Stimulating Factor-2 Receptor, Beta, Low-Affinity, GM-CSF/IL-3/IL-5 Receptor Common Beta-Chain, Cytokine Receptor Common Subunit Beta, CD131 Antigen, CD131 Description GM-CSF Receptor-beta, also known as CSF2RB is a member of the type I cytokine receptor family. CSF2RB is a high affinity receptor for interleukin-3, interleukin-5 as well as granulocyte- macrophage colony-stimulating factor. The CSF2RB unique form of receptor assembly applies also to IL-3 and IL-5 receptors, providing a structural basis for understanding their activation mechanism which is essential for the development of therapeutics. Human recombinant GM-CSF Receptor-beta produced in Sf9 cells is a single, glycosylated polypeptide chain of 435 amino acids (17-443 a.a), having a molecular mass of 49.7 kDa (migrates at 40-57 kDa on SDS-PAGE under reducing conditions). CSF2RB is fused to an 8 amino acid His-tag at the C-terminus and purified using proprietary chromatographic techniques. Quantity 10 µg Molecular Mass 49.7 kDa Source Sf9 insect cells Biological-Activity NA Specific Activity NA Formulation Sterile-filtered, clear protein solution (0.5 mg/ml) containing phosphate-buffered saline (pH 7.4) and 10% glycerol. Reconstitution Please Note: Always centrifuge product briefly before opening vial. -

Cd4/Cd8 Panel, Blood
Lab Dept: Flow and Immunology Test Name: CD4/CD8 PANEL, BLOOD General Information Lab Order Codes: C48P Synonyms: Helper/Suppressor Ratio; T- cells; T-cell subsets; T-cell phenotyping See also: Immune Status Panel and Comprehensive Immune Status Panel CPT Codes: 86359 – T cells, total count 86360 – T cells; absolute CD4 and CD8 count, including ratio Test Includes: CD4(CD3+) and CD8(CD3+) relative percentages, absolute values and a calculated Helper/Suppressor ratio. Logistics Test Indications: This is a minimal antibody panel to monitor immune status. Lab Testing Sections: Flow Cytometry Phone Numbers: MIN Lab: 612-813-6280 STP Lab: 651-220-6550 Test Availability: 3 times weekly determined by volume. Transport collected specimen immediately to Flow Cytometry. Routine testing is not available on weekends or holidays. Therefore, specimens cannot be used if drawn the day before a 3 day weekend such as Memorial Day, Labor Day or major holiday that falls on a Monday or Friday. Turnaround Time: 1 – 3 days Special Instructions: See Test Availability Specimen Specimen Type: Whole blood Container: Lavender top (EDTA) tube Draw Volume: 2 mL blood in a 2 mL Lavender (EDTA) tube Minimum volume: 0.5 mL in an EDTA microtainer Collection: Routine venipuncture Special Processing: Keep sample at room temperature and forward promptly to the laboratory. Do not centrifuge, refrigerate, or freeze sample. Patient Preparation: None Sample Rejection: Specimens will not be processed that are clotted; hemolyzed; greater than 72 hours old; collected in the wrong tube type (0.5 mL in a 2 mL tube), or that have been held or handled at a temperature other than room temperature Interpretive Reference Range: Age-dependant reference ranges provided with results. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -
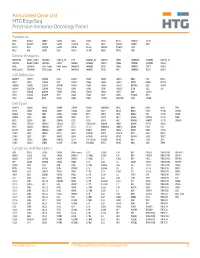
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2