GM-CSF Receptor-Beta, Human, Recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor Beta
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Mechanism of Homodimeric Cytokine Receptor Activation and Dysregulation by Oncogenic Mutations
This is a repository copy of Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. White Rose Research Online URL for this paper: https://eprints.whiterose.ac.uk/155270/ Version: Accepted Version Article: Wilmes, Stephan, Hafer, Maximillian, Vuorio, Joni et al. (15 more authors) (2020) Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science. pp. 643-652. ISSN 0036-8075 https://doi.org/10.1126/science.aaw3242 Reuse Items deposited in White Rose Research Online are protected by copyright, with all rights reserved unless indicated otherwise. They may be downloaded and/or printed for private study, or other acts as permitted by national copyright laws. The publisher or other rights holders may allow further reproduction and re-use of the full text version. This is indicated by the licence information on the White Rose Research Online record for the item. Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Submitted Manuscript: Confidential Title: Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations Authors: 5 Stephan Wilmes1, 2*, Maximillian Hafer1*, Joni Vuorio3,4, Julie A. Tucker5, Hauke Winkelmann1, Sara Löchte1, Tess A. Stanly5, Katiuska D. Pulgar Prieto5, Chetan Poojari3, Vivek Sharma3,6, Christian P. Richter1, Rainer Kurre1, Stevan R. Hubbard7, K. Christopher Garcia8,9, Ignacio Moraga2, Ilpo Vattulainen3,4,10†, Ian S. Hitchcock5† and Jacob Piehler1† Affiliations: 10 1 Department of Biology and Center of Cellular Nanoanalytics, University of Osnabrück, 49076 Osnabrück, Germany. -

Stony Brook University
SSStttooonnnyyy BBBrrrooooookkk UUUnnniiivvveeerrrsssiiitttyyy The official electronic file of this thesis or dissertation is maintained by the University Libraries on behalf of The Graduate School at Stony Brook University. ©©© AAAllllll RRRiiiggghhhtttsss RRReeessseeerrrvvveeeddd bbbyyy AAAuuuttthhhooorrr... Regulation of Dimerization and Activation of the Thrombopoietin Receptor A Dissertation Presented by Miki Itaya to The Graduate School in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Biochemistry and Structural Biology Stony Brook University December 2012 Copyright by Miki Itaya 2012 Stony Brook University The Graduate School Miki Itaya We, the dissertation committee for the above candidate for the Doctor of Philosophy degree, hereby recommend acceptance of this dissertation. Steven O. Smith, Ph.D. - Dissertation Advisor Professor, Department of Biochemistry and Cell Biology Erwin London, Ph.D. - Chairperson of Defense Professor, Department of Biochemistry and Cell Biology Robert C. Rizzo, Ph.D. Associate Professor, Department of Applied Mathematics and Statistics Nancy Reich Marshall, Ph.D. Professor, Department of Molecular Genetics and Microbiology This dissertation is accepted by the Graduate School Charles Taber Interim Dean of the Graduate School ii Abstract of the Dissertation Regulation of Dimerization and Activation of the Thrombopoietin Receptor by Miki Itaya Doctor of Philosophy in Biochemistry and Structural Biology Stony Brook University 2012 The thrombopoietin receptor (TpoR) is -

Immuno-Endocrine Interactions in Intestinal Inflammation
IMMUNO-ENDOCRINE INTERACTIONS IN INTESTINAL INFLAMMATION PhD Thesis- Shajib, MS; McMaster University-Medical Sciences Immuno-endocrine interactions in intestinal inflammation By Md. Sharif Shajib, BSc. (Hons) A Thesis Submitted to the School of Graduate Studies in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy McMaster University © Copyright by Md. Sharif Shajib, 2017. PhD Thesis- Shajib, MS; McMaster University-Medical Sciences Descriptive notes Doctor of Philosophy (2017) McMaster University, Hamilton, Ontario (Medical Sciences) TITLE Immuno-endocrine interactions in intestinal inflammation AUTHOR Md. Sharif Shajib, BSc. (Hons) SUPERVISOR Dr. Waliul I. Khan NUMBER OF PAGES: XX, 292. II PhD Thesis- Shajib, MS; McMaster University-Medical Sciences Lay abstract The gut produces most of the serotonin found in our body, where it regulates many normal functions. A group of special cells, named enterochromaffin cells, produces nearly all of the serotonin in the gut. In diseases of the gut, especially ones that involve inflammation resulting in symptoms like abdominal pain, diarrhea and bleeding, the number of these cells and serotonin concentration are different from that in the normal gut. I found that these changes are controlled by a particular protein produced by immune cells, called interleukin-13, and alteration in serotonin levels, in turn, contributes to the inflammatory process. Our laboratory experiments with cells and animals establish this connection between interleukin-13 and serotonin in gut inflammation. We further confirm this association between interleukin-13 and serotonin in human inflammatory bowel disease. Moreover, we identify a potential genetic cause of these changes in serotonin concentrations which may ultimately result in inflammatory bowel disease. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -
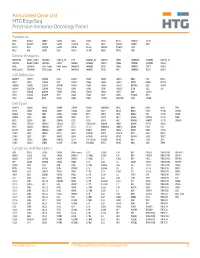
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2 -

Facteurs Génétiques De Prédisposition a La Maladie Coeliaque Et L’Oesophagite Éosinophilique
Université de Montréal FACTEURS GÉNÉTIQUES DE PRÉDISPOSITION A LA MALADIE COELIAQUE ET L’OESOPHAGITE ÉOSINOPHILIQUE Par Freha Nour el Hayet CHERIEF Département de Microbiologie et Immunologie Faculté de Médecine Mémoire présenté à la Faculté des études supérieures en vue de l’obtention du grade de Maîtrise en Microbiologie et Immunologie Novembre 2012 © Freha Nour el Hayet CHERIEF, 2012 i Université de Montréal Faculté des études supérieures Ce mémoire intitulé : FACTEURS GÉNÉTIQUES DE PRÉDISPOSITION A LA MALADIE COELIAQUE ET L’OESOPHAGITE ÉOSINOPHILIQUE Présenté par : Freha nour el hayet CHERIEF a été évalué par un jury composé des personnes suivantes : Ali Ahmad Président-rapporteur Idriss Djilali-Saïah Directeur de recherche Cristophe Faure Membre du jury ii Résumé Les maladies immunitaires chroniques incluant les maladies auto-immunes et inflammatoires touchent 20 à 25% de la population des pays occidentaux. La comparaison des taux de concordance chez les jumeaux ou l’histoire familiale de sujets atteints de la maladie cœliaque (maladie auto-immune de l’intestin) ou de l’œsophagite éosinophilique (maladie inflammatoire de l’œsophage) indiquent que des facteurs génétiques et environnementaux interviennent dans la susceptibilité à ces maladies. Cependant, ces études ne distinguent pas de manière claire la prédisposition génétique selon l’hétérogénéité clinique (enfants versus adultes) ou ethnique (stratification des populations). Méthodes. Les haplotypes HLA de prédisposition à la maladie cœliaque et les polymorphismes des gènes candidats -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

WO 2018/067991 Al 12 April 2018 (12.04.2018) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/067991 Al 12 April 2018 (12.04.2018) W !P O PCT (51) International Patent Classification: achusetts 021 15 (US). THE BROAD INSTITUTE, A61K 51/10 (2006.01) G01N 33/574 (2006.01) INC. [US/US]; 415 Main Street, Cambridge, Massachu C07K 14/705 (2006.01) A61K 47/68 (2017.01) setts 02142 (US). MASSACHUSETTS INSTITUTE OF G01N 33/53 (2006.01) TECHNOLOGY [US/US]; 77 Massachusetts Avenue, Cambridge, Massachusetts 02139 (US). (21) International Application Number: PCT/US2017/055625 (72) Inventors; and (71) Applicants: KUCHROO, Vijay K. [IN/US]; 30 Fairhaven (22) International Filing Date: Road, Newton, Massachusetts 02149 (US). ANDERSON, 06 October 2017 (06.10.2017) Ana Carrizosa [US/US]; 110 Cypress Street, Brookline, (25) Filing Language: English Massachusetts 02445 (US). MADI, Asaf [US/US]; c/o The Brigham and Women's Hospital, Inc., 75 Francis (26) Publication Language: English Street, Boston, Massachusetts 021 15 (US). CHIHARA, (30) Priority Data: Norio [US/US]; c/o The Brigham and Women's Hospital, 62/405,835 07 October 2016 (07.10.2016) US Inc., 75 Francis Street, Boston, Massachusetts 021 15 (US). REGEV, Aviv [US/US]; 15a Ellsworth Ave, Cambridge, (71) Applicants: THE BRIGHAM AND WOMEN'S HOSPI¬ Massachusetts 02139 (US). SINGER, Meromit [US/US]; TAL, INC. [US/US]; 75 Francis Street, Boston, Mass c/o The Broad Institute, Inc., 415 Main Street, Cambridge, (54) Title: MODULATION OF NOVEL IMMUNE CHECKPOINT TARGETS CD4 FIG. -

Conserved Transcriptomic Profile Between Mouse and Human Colitis
ARTICLE https://doi.org/10.1038/s41467-019-10769-x OPEN Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification Paulo Czarnewski1, Sara M. Parigi1, Chiara Sorini 1, Oscar E. Diaz 1, Srustidhar Das1, Nicola Gagliani1,2,3 & Eduardo J. Villablanca 1,3 1234567890():,; Clinical manifestations and response to therapies in ulcerative colitis (UC) are hetero- geneous, yet patient classification criteria for tailored therapies are currently lacking. Here, we present an unsupervised molecular classification of UC patients, concordant with response to therapy in independent retrospective cohorts. We show that classical clustering of UC patient tissue transcriptomic data sets does not identify clinically relevant profiles, likely due to associated covariates. To overcome this, we compare cross-sectional human data sets with a newly generated longitudinal transcriptome profile of murine DSS-induced colitis. We show that the majority of colitis risk-associated gene expression peaks during the inflammatory rather than the recovery phase. Moreover, we achieve UC patient clustering into two distinct transcriptomic profiles, differing in neutrophil-related gene activation. Notably, 87% of patients in UC1 cluster are unresponsive to two most widely used biological therapies. These results demonstrate that cross-species comparison enables stratification of patients undistinguishable by other molecular approaches. 1 Immunology and Allergy Unit, Department of Medicine, Solna, Karolinska Institute and University Hospital, 17176 Stockholm, Sweden. 2 Department of Medicine and Department of General, Visceral and Thoracic Surgery, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany. 3These authors jointly supervised this work: Nicola Gagliani, Eduardo J. Villablanca. Correspondence and requests for materials should be addressed to E.J.V. -

A Closer Look at JAK/STAT Signaling Pathway Emira Bousoik Chapman University
Chapman University Chapman University Digital Commons Pharmacy Faculty Articles and Research School of Pharmacy 7-31-2018 “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway Emira Bousoik Chapman University Hamidreza Montazeri Aliabadi Chapman University, [email protected] Follow this and additional works at: https://digitalcommons.chapman.edu/pharmacy_articles Part of the Amino Acids, Peptides, and Proteins Commons, Cancer Biology Commons, Cell Anatomy Commons, Cell Biology Commons, Enzymes and Coenzymes Commons, Oncology Commons, and the Other Pharmacy and Pharmaceutical Sciences Commons Recommended Citation Bousoik E, Montazeri Aliabadi H. “Do we know jack” about JAK? A closer look at JAK/STAT signaling pathway. Front. Oncol. 2018;8:287. doi: 10.3389/fonc.2018.00287 This Article is brought to you for free and open access by the School of Pharmacy at Chapman University Digital Commons. It has been accepted for inclusion in Pharmacy Faculty Articles and Research by an authorized administrator of Chapman University Digital Commons. For more information, please contact [email protected]. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway Comments This article was originally published in Frontiers in Oncology, volume 8, in 2018. DOI: 10.3389/ fonc.2018.00287 Creative Commons License This work is licensed under a Creative Commons Attribution 4.0 License. Copyright The uthora s This article is available at Chapman University Digital Commons: https://digitalcommons.chapman.edu/pharmacy_articles/590