Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L
Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L. Gommerman This information is current as J Immunol 2020; 204:1075-1083; ; of September 28, 2021. doi: 10.4049/jimmunol.1900710 http://www.jimmunol.org/content/204/5/1075 Downloaded from References This article cites 152 articles, 56 of which you can access for free at: http://www.jimmunol.org/content/204/5/1075.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 28, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2020 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen, and Jennifer L. Gommerman The mammalian intestine is a complex environment shaping the ensuing immune response. In this Brief Review,we that is constantly exposed to Ags derived from food, review the different types of classical DC (cDC) located in the microbiota, and metabolites. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Dendritic Cell Progenitor Trafficking and Identification and Functional Analyses of Dendritic Cells with Distinct Developmental Origin
Dissertation zum Erwerb des Doctor of Philosophy (Ph.D.) an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München Dendritic cell progenitor trafficking and identification and functional analyses of dendritic cells with distinct developmental origin vorgelegt von: Johanna Marie Salvermoser aus: Pfaffenhofen an der Ilm Jahr: 2020 _____________________ First supervisor: Prof. Dr. Barbara Schraml Second supervisor: Prof. Dr. Anne Krug Third supervisor: Prof. Dr. Christian Schulz Fourth supervisor: PD. Dr. rer. nat. Caspar Ohnmacht Dean: Prof. Dr. med. dent. Reinhard Hickel Datum der Verteidigung: 09.03.2020 2 Abstract ABSTRACT Conventional dendritic cells (cDCs), are the major antigen-presenting cell type that bridges the innate and adaptive immune system. DCs are constantly replenished from myeloid bone marrow progenitors which latest stage, pre-cDC, leave the BM, seeds the peripheral tissues and further differentiates into two functionally and developmentally distinct subsets, cDC1 and cDC2. This study aimed to investigate DC development by assessing the trafficking of pre-cDCs and by analyzing the effect of a specific depletion of DC progenitors. The signals that regulate the recruitment of pre-cDCs to different peripheral organs are poorly understood. Therefore, this study aimed to identify pre-cDCs in different peripheral organs and to find differences in expression pattern of trafficking receptors. In this study 39 trafficking receptors have been identified to be expressed on pre-cDCs of the analysed tissues and showed differences in the expression patterns between peripheral organs. These receptors are interesting candidates to further study differences in the recruitment of pre-cDCs to different peripheral tissues This can provide possibilities to influence the recruitment of pre-cDCs in certain diseases, where the replenishment of cDCs is accelerated. -

Targeting Dendritic Cells with Antigen-Delivering Antibodies for Amelioration of Autoimmunity in Animal Models of Multiple Sclerosis and Other Autoimmune Diseases
antibodies Review Targeting Dendritic Cells with Antigen-Delivering Antibodies for Amelioration of Autoimmunity in Animal Models of Multiple Sclerosis and Other Autoimmune Diseases Courtney A. Iberg and Daniel Hawiger * Department of Molecular Microbiology and Immunology, Saint Louis University School of Medicine, Doisy Research Center, 1205 Carr Lane, St. Louis, MO 63104, USA; [email protected] * Correspondence: [email protected] Received: 31 March 2020; Accepted: 30 April 2020; Published: 15 June 2020 Abstract: The specific targeting of dendritic cells (DCs) using antigen-delivering antibodies has been established to be a highly efficient protocol for the induction of tolerance and protection from autoimmune processes in experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis (MS), as well as in some other animal disease models. As the specific mechanisms of such induced tolerance are being investigated, the newly gained insights may also possibly help to design effective treatments for patients. Here we review approaches applied for the amelioration of autoimmunity in animal models based on antibody-mediated targeting of self-antigens to DCs. Further, we discuss relevant mechanisms of immunological tolerance that underlie such approaches, and we also offer some future perspectives for the application of similar methods in certain related disease settings such as transplantation. Keywords: dendritic cells; tolerance; antigen targeting; chimeric antibodies; autoimmunity; multiple sclerosis; diabetes 1. Introduction Over one hundred years ago, Paul Ehrlich coined the term “horror autotoxicus” to define an immune attack against an organism’s healthy tissues [1]. Since then, our knowledge of the complex mechanisms of the immune system as well as our understanding of the pathogenesis of specific autoimmune diseases have grown tremendously. -

Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor-Infiltrating Lymphocytes
Published OnlineFirst February 18, 2016; DOI: 10.1158/0008-5472.CAN-15-1548 Cancer Tumor and Stem Cell Biology Research Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor- Infiltrating Lymphocytes Geoffrey J. Markowitz1, Pengyuan Yang1,2,3, Jing Fu3, Gregory A. Michelotti4, Rui Chen1, Jianhua Sui5, Bin Yang2, Wen-Hao Qin3, Zheng Zhang6, Fu-Sheng Wang6, Anna Mae Diehl4, Qi-Jing Li7, Hongyang Wang3, and Xiao-Fan Wang1 Abstract Chronic inflammation in liver tissue is an underlying cause of IL18R1 deletion increased tumor burden. Mechanistically, we hepatocellular carcinoma. High levels of inflammatory cytokine foundthatIL18exertedinflammation-dependent tumor-sup- IL18 in the circulation of patients with hepatocellular carcinoma pressive effects largely by promoting the differentiation, activ- correlates with poor prognosis. However, conflicting results have ity, and survival of tumor-infiltrating T cells. Finally, differences been reported for IL18 in hepatocellular carcinoma development in the expression of IL18 in tumor tissue versus nontumor and progression. In this study, we used tissue specimens from tissueweremorepredictiveofpatientoutcomethanoverall hepatocellular carcinoma patients and clinically relevant mouse tissue expression. Taken together, our findings resolve a long- models of hepatocellular carcinoma to evaluate IL18 expression standing contradiction regarding a tumor-suppressive role for and function. In a mouse model of liver fibrosis that recapitulates IL18 in established hepatocellular carcinoma and provide a a tumor-promoting microenvironment, global deletion of the mechanistic explanation for the complex relationship between IL18 receptor IL18R1 enhanced tumor growth and burden. Sim- its expression pattern and hepatocellular carcinoma prognosis. ilarly, in a carcinogen-induced model of liver tumorigenesis, Cancer Res; 76(8); 1–12. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -
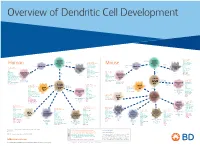
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -
GM-CSF Receptor-Beta, Human, Recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor Beta
GM-CSF Receptor-beta, human, recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor beta Instruction Manual Catalog Number C-60430 Synonyms CSF2RB,Colony Stimulating Factor 2 Receptor, Beta, Low-Affinity (Granulocyte-Macrophage), GM-CSF/IL-3/IL-5 Receptor Common Beta Subunit, CDw131, IL3RB, SMDP5, IL5RB, Interleukin 3 Receptor/Granulocyte-Macrophage Colony Stimulating Factor 3 Receptor, Beta (High Affinity), Colony-Stimulating Factor-2 Receptor, Beta, Low-Affinity, GM-CSF/IL-3/IL-5 Receptor Common Beta-Chain, Cytokine Receptor Common Subunit Beta, CD131 Antigen, CD131 Description GM-CSF Receptor-beta, also known as CSF2RB is a member of the type I cytokine receptor family. CSF2RB is a high affinity receptor for interleukin-3, interleukin-5 as well as granulocyte- macrophage colony-stimulating factor. The CSF2RB unique form of receptor assembly applies also to IL-3 and IL-5 receptors, providing a structural basis for understanding their activation mechanism which is essential for the development of therapeutics. Human recombinant GM-CSF Receptor-beta produced in Sf9 cells is a single, glycosylated polypeptide chain of 435 amino acids (17-443 a.a), having a molecular mass of 49.7 kDa (migrates at 40-57 kDa on SDS-PAGE under reducing conditions). CSF2RB is fused to an 8 amino acid His-tag at the C-terminus and purified using proprietary chromatographic techniques. Quantity 10 µg Molecular Mass 49.7 kDa Source Sf9 insect cells Biological-Activity NA Specific Activity NA Formulation Sterile-filtered, clear protein solution (0.5 mg/ml) containing phosphate-buffered saline (pH 7.4) and 10% glycerol. Reconstitution Please Note: Always centrifuge product briefly before opening vial. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -

Development of Autoimmune Hair Loss Disease Alopecia Areata Is Associated with Cardiac Dysfunction in C3H/ Hej Mice
Development of Autoimmune Hair Loss Disease Alopecia Areata Is Associated with Cardiac Dysfunction in C3H/ HeJ Mice Eddy Wang1, Katy Chong2, Mei Yu1, Noushin Akhoundsadegh1, David J. Granville3, Jerry Shapiro4, Kevin J. McElwee1* 1 Department of Dermatology and Skin Science, University of British Columbia, Vancouver, BC, Canada, 2 University of British Columbia, Vancouver, BC, Canada, 3 Department of Pathology and Laboratory Medicine, James Hogg Research Centre, Institute for Heart and Lung Health, University of British Columbia, Vancouver, BC, Canada, 4 Department of Dermatology and Skin Science, Vancouver General Hospital, Vancouver, BC, Canada Abstract Alopecia areata (AA) is a chronic autoimmune hair loss disease that affects several million men, women and children worldwide. Previous studies have suggested a link between autoimmunity, stress hormones, and increased cardiovascular disease risk. In the current study, histology, immunohistology, quantitative PCR (qPCR) and ELISAs were used to assess heart health in the C3H/HeJ mouse model for AA and heart tissue response to adrenocorticotropic hormone (ACTH) exposure. Mice with AA exhibited both atrial and ventricular hypertrophy, and increased collagen deposition compared to normal- haired littermates. QPCR revealed significant increases in Il18 (4.6-fold), IL18 receptor-1 (Il18r1; 2.8-fold) and IL18 binding protein (Il18bp; 5.2-fold) in AA hearts. Time course studies revealed a trend towards decreased Il18 in acute AA compared to controls while Il18r1, Il18bp and Casp1 showed similar trends to those of chronic AA affected mice. Immunohistochemistry showed localization of IL18 in chronic AA mouse atria. ELISA indicated cardiac troponin-I (cTnI) was elevated in the serum and significantly increased in AA heart tissue.