Targeting Dendritic Cells with Antigen-Delivering Antibodies for Amelioration of Autoimmunity in Animal Models of Multiple Sclerosis and Other Autoimmune Diseases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
IFM Innate Immunity Infographic
UNDERSTANDING INNATE IMMUNITY INTRODUCTION The immune system is comprised of two arms that work together to protect the body – the innate and adaptive immune systems. INNATE ADAPTIVE γδ T Cell Dendritic B Cell Cell Macrophage Antibodies Natural Killer Lymphocites Neutrophil T Cell CD4+ CD8+ T Cell T Cell TIME 6 hours 12 hours 1 week INNATE IMMUNITY ADAPTIVE IMMUNITY Innate immunity is the body’s first The adaptive, or acquired, immune line of immunological response system is activated when the innate and reacts quickly to anything that immune system is not able to fully should not be present. address a threat, but responses are slow, taking up to a week to fully respond. Pathogen evades the innate Dendritic immune system T Cell Cell Through antigen Pathogen presentation, the dendritic cell informs T cells of the pathogen, which informs Macrophage B cells B Cell B cells create antibodies against the pathogen Macrophages engulf and destroy Antibodies label invading pathogens pathogens for destruction Scientists estimate innate immunity comprises approximately: The adaptive immune system develops of the immune memory of pathogen exposures, so that 80% system B and T cells can respond quickly to eliminate repeat invaders. IMMUNE SYSTEM AND DISEASE If the immune system consistently under-responds or over-responds, serious diseases can result. CANCER INFLAMMATION Innate system is TOO ACTIVE Innate system NOT ACTIVE ENOUGH Cancers grow and spread when tumor Certain diseases trigger the innate cells evade detection by the immune immune system to unnecessarily system. The innate immune system is respond and cause excessive inflammation. responsible for detecting cancer cells and This type of chronic inflammation is signaling to the adaptive immune system associated with autoimmune and for the destruction of the cancer cells. -

Adoptive Cell Therapy Using Engineered Natural Killer Cells
Bone Marrow Transplantation (2019) 54:785–788 https://doi.org/10.1038/s41409-019-0601-6 REVIEW ARTICLE Adoptive cell therapy using engineered natural killer cells Katayoun Rezvani1 © The Author(s), under exclusive licence to Springer Nature Limited 2019 Abstract The generation of autologous T cells expressing a chimeric antigen receptor (CAR) have revolutionized the field of adoptive cellular therapy. CAR-T cells directed against CD19 have resulted in remarkable clinical responses in patients affected by B-lymphoid malignancies. However, the production of allogeneic CAR-T cells products remains expensive and clinically challenging. Moreover, the toxicity profile of CAR T-cells means that currently these life-saving treatments are only delivered in specialized centers. Therefore, efforts are underway to develop reliable off-the-shelf cellular products with acceptable safety profiles for the treatment of patients with cancer. Natural killer (NK) cells are innate effector lymphocytes with potent antitumor activity. The availability of NK cells from multiple sources and their proven safety profile in the allogeneic setting positions them as attractive contenders for cancer immunotherapy. In this review, we discuss advantages and potential drawbacks of using NK cells as a novel cellular therapy against hematologic malignancies, as well as strategies 1234567890();,: 1234567890();,: to further enhance their effector function. Introduction need for inpatient care may ultimately be economically unviable for many health care systems; (iii) the longer time Adoptive cell therapy has become a powerful treatment that is required to generate CAR T-cells may result in modality for advanced cancers refractory to conventional unavoidable delays in therapy, especially for patients with therapy. -

Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L
Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen and Jennifer L. Gommerman This information is current as J Immunol 2020; 204:1075-1083; ; of September 28, 2021. doi: 10.4049/jimmunol.1900710 http://www.jimmunol.org/content/204/5/1075 Downloaded from References This article cites 152 articles, 56 of which you can access for free at: http://www.jimmunol.org/content/204/5/1075.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 28, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2020 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Dendritic Cell Subsets in Intestinal Immunity and Inflammation Tian Sun, Albert Nguyen, and Jennifer L. Gommerman The mammalian intestine is a complex environment shaping the ensuing immune response. In this Brief Review,we that is constantly exposed to Ags derived from food, review the different types of classical DC (cDC) located in the microbiota, and metabolites. -

Dendritic Cell Progenitor Trafficking and Identification and Functional Analyses of Dendritic Cells with Distinct Developmental Origin
Dissertation zum Erwerb des Doctor of Philosophy (Ph.D.) an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München Dendritic cell progenitor trafficking and identification and functional analyses of dendritic cells with distinct developmental origin vorgelegt von: Johanna Marie Salvermoser aus: Pfaffenhofen an der Ilm Jahr: 2020 _____________________ First supervisor: Prof. Dr. Barbara Schraml Second supervisor: Prof. Dr. Anne Krug Third supervisor: Prof. Dr. Christian Schulz Fourth supervisor: PD. Dr. rer. nat. Caspar Ohnmacht Dean: Prof. Dr. med. dent. Reinhard Hickel Datum der Verteidigung: 09.03.2020 2 Abstract ABSTRACT Conventional dendritic cells (cDCs), are the major antigen-presenting cell type that bridges the innate and adaptive immune system. DCs are constantly replenished from myeloid bone marrow progenitors which latest stage, pre-cDC, leave the BM, seeds the peripheral tissues and further differentiates into two functionally and developmentally distinct subsets, cDC1 and cDC2. This study aimed to investigate DC development by assessing the trafficking of pre-cDCs and by analyzing the effect of a specific depletion of DC progenitors. The signals that regulate the recruitment of pre-cDCs to different peripheral organs are poorly understood. Therefore, this study aimed to identify pre-cDCs in different peripheral organs and to find differences in expression pattern of trafficking receptors. In this study 39 trafficking receptors have been identified to be expressed on pre-cDCs of the analysed tissues and showed differences in the expression patterns between peripheral organs. These receptors are interesting candidates to further study differences in the recruitment of pre-cDCs to different peripheral tissues This can provide possibilities to influence the recruitment of pre-cDCs in certain diseases, where the replenishment of cDCs is accelerated. -

Natural Killer Cells Associated with SARS-Cov-2 Viral RNA Shedding
Bao et al. Exp Hematol Oncol (2021) 10:5 https://doi.org/10.1186/s40164-021-00199-1 Experimental Hematology & Oncology LETTER TO THE EDITOR Open Access Natural killer cells associated with SARS-CoV-2 viral RNA shedding, antibody response and mortality in COVID-19 patients Changqian Bao1†, Xiandong Tao2,3†, Wei Cui4†, Yuanyuan Hao1, Shuaike Zheng5, Bin Yi2,3, Tiewen Pan2, Ken H. Young6* and Wenbin Qian1,7* Abstract Coronavirus disease 2019 (COVID-19) is a novel infectious viral disease caused by the severe acute respiratory syn- drome coronavirus 2 (SARS-CoV-2). Two consecutively negative SARS-CoV-2 viral RNA test ( interval 24 hours), improved respiratory symptoms and obvious absorption of infammation in pulmonary imaging are≥ the discharge criteria for COVID-19 patients. The clearance profle of viral RNA in the upper respiratory tract specimens, including nasopharyngeal swab and/or oropharyngeal swabs, is related to innate immune cells such as Natural Killer cells. A total of 168 patients were included for the study. In this cohort, non-severe and severe groups showed signifcant dif- ferences in white blood cells, neutrophils, lymphocytes, basophils and platelets counts, as well as in infection related parameters such as CRP and serum cytokine IL-6. For lymphocyte subsets tests at admission, the severe group dis- played signifcantly lower cell counts than the non-severe group. Higher counts of total T cells, CD4 T cells, CD8 T cells, and NK cells in peripheral blood showed a signifcant correlation with the shorter time taken to+ obtain the frst+ negative viral RNA test and frst positive IgM/ IgG antibody test. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

Adaptive Immune Systems
Immunology 101 (for the Non-Immunologist) Abhinav Deol, MD Assistant Professor of Oncology Wayne State University/ Karmanos Cancer Institute, Detroit MI Presentation originally prepared and presented by Stephen Shiao MD, PhD Department of Radiation Oncology Cedars-Sinai Medical Center Disclosures Bristol-Myers Squibb – Contracted Research What is the immune system? A network of proteins, cells, tissues and organs all coordinated for one purpose: to defend one organism from another It is an infinitely adaptable system to combat the complex and endless variety of pathogens it must address Outline Structure of the immune system Anatomy of an immune response Role of the immune system in disease: infection, cancer and autoimmunity Organs of the Immune System Major organs of the immune system 1. Bone marrow – production of immune cells 2. Thymus – education of immune cells 3. Lymph Nodes – where an immune response is produced 4. Spleen – dual role for immune responses (especially antibody production) and cell recycling Origins of the Immune System B-Cell B-Cell Self-Renewing Common Progenitor Natural Killer Lymphoid Cell Progenitor Thymic T-Cell Selection Hematopoetic T-Cell Stem Cell Progenitor Dendritic Cell Myeloid Progenitor Granulocyte/M Macrophage onocyte Progenitor The Immune Response: The Art of War “Know your enemy and know yourself and you can fight a hundred battles without disaster.” -Sun Tzu, The Art of War Immunity: Two Systems and Their Key Players Adaptive Immunity Innate Immunity Dendritic cells (DC) B cells Phagocytes (Macrophages, Neutrophils) Natural Killer (NK) Cells T cells Dendritic Cells: “Commanders-in-Chief” • Function: Serve as the gateway between the innate and adaptive immune systems. -
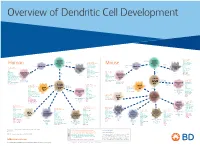
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Eosinophil Response Against Classical and Emerging
REVIEWS Eosinophil Response Against Classical and Emerging Respiratory Viruses: COVID-19 Rodrigo-Muñoz JM1,2, Sastre B1,2, Cañas JA1,2, Gil-Martínez M1, Redondo N1, del Pozo V1,2 1Immunology Department, Instituto de Investigación Sanitaria (IIS) Fundación Jiménez Díaz, Madrid, Spain 2CIBER de Enfermedades Respiratorias (CIBERES), Madrid, Spain J Investig Allergol Clin Immunol 2021; Vol. 31(2): 94-107 doi: 10.18176/jiaci.0624 Abstract Eosinophils were discovered more than 140 years ago. These polymorphonuclear leukocytes have a very active metabolism and contain numerous intracellular secretory granules that enable multiple effects on both health and disease status. Classically, eosinophils have been considered important immune cells in the pathogenesis of inflammatory processes (eg, parasitic helminth infections) and allergic or pulmonary diseases (eg, asthma) and are always associated with a type 2 immune response. Furthermore, in recent years, eosinophils have been linked to the immune response by conferring host protection against fungi, bacteria, and viruses, which they recognize through several molecules, such as toll-like receptors and the retinoic acid–inducible gene 1–like receptor. The immune protection provided by eosinophils is exerted through multiple mechanisms and properties. Eosinophils contain numerous cytoplasmatic granules that release cationic proteins, cytokines, chemokines, and other molecules, all of which contribute to their functioning. In addition to the competence of eosinophils as effector cells, their capabilities as antigen-presenting cells enable them to act in multiple situations, thus promoting diverse aspects of the immune response. This review summarizes various aspects of eosinophil biology, with emphasis on the mechanisms used and roles played by eosinophils in host defence against viral infections and response to vaccines. -
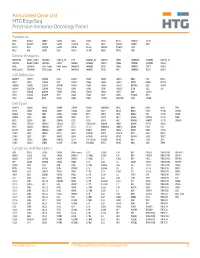
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2 -

Understanding the Immune System: How It Works
Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute NIH Publication No. 03-5423 September 2003 www.niaid.nih.gov www.nci.nih.gov Contents 1 Introduction 2 Self and Nonself 3 The Structure of the Immune System 7 Immune Cells and Their Products 19 Mounting an Immune Response 24 Immunity: Natural and Acquired 28 Disorders of the Immune System 34 Immunology and Transplants 36 Immunity and Cancer 39 The Immune System and the Nervous System 40 Frontiers in Immunology 45 Summary 47 Glossary Introduction he immune system is a network of Tcells, tissues*, and organs that work together to defend the body against attacks by “foreign” invaders. These are primarily microbes (germs)—tiny, infection-causing Bacteria: organisms such as bacteria, viruses, streptococci parasites, and fungi. Because the human body provides an ideal environment for many microbes, they try to break in. It is the immune system’s job to keep them out or, failing that, to seek out and destroy them. Virus: When the immune system hits the wrong herpes virus target or is crippled, however, it can unleash a torrent of diseases, including allergy, arthritis, or AIDS. The immune system is amazingly complex. It can recognize and remember millions of Parasite: different enemies, and it can produce schistosome secretions and cells to match up with and wipe out each one of them.