Conserved Transcriptomic Profile Between Mouse and Human Colitis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Cancer-Associated Fibroblasts Promote Prostate Tumor Growth And
Neuwirt et al. Cell Communication and Signaling (2020) 18:11 https://doi.org/10.1186/s12964-019-0505-5 RESEARCH Open Access Cancer-associated fibroblasts promote prostate tumor growth and progression through upregulation of cholesterol and steroid biosynthesis Hannes Neuwirt1, Jan Bouchal2, Gvantsa Kharaishvili2, Christian Ploner3, Karin Jöhrer4,5, Florian Pitterl6, Anja Weber7, Helmut Klocker7 and Iris E. Eder7* Abstract Background: Androgen receptor targeted therapies have emerged as an effective tool to manage advanced prostate cancer (PCa). Nevertheless, frequent occurrence of therapy resistance represents a major challenge in the clinical management of patients, also because the molecular mechanisms behind therapy resistance are not yet fully understood. In the present study, we therefore aimed to identify novel targets to intervene with therapy resistance using gene expression analysis of PCa co-culture spheroids where PCa cells are grown in the presence of cancer-associated fibroblasts (CAFs) and which have been previously shown to be a reliable model for antiandrogen resistance. Methods: Gene expression changes of co-culture spheroids (LNCaP and DuCaP seeded together with CAFs) were identified by Illumina microarray profiling. Real-time PCR, Western blotting, immunohistochemistry and cell viability assays in 2D and 3D culture were performed to validate the expression of selected targets in vitro and in vivo. Cytokine profiling was conducted to analyze CAF-conditioned medium. Results: Gene expression analysis of co-culture spheroids revealed that CAFs induced a significant upregulation of cholesterol and steroid biosynthesis pathways in PCa cells. Cytokine profiling revealed high amounts of pro- inflammatory, pro-migratory and pro-angiogenic factors in the CAF supernatant. In particular, two genes, 3-hydroxy- 3-methylglutaryl-Coenzyme A synthase 2 (HMGCS2) and aldo-keto reductase family 1 member C3 (AKR1C3), were significantly upregulated in PCa cells upon co-culture with CAFs. -

Intra- and Inter-Cellular Rewiring of the Human Colon During Ulcerative Colitis
Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Smillie, Christopher S. et al. “Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis.” Cell, 178, 3 (July 2019): 714–730.e22 © 2019 The Author(s) As Published 10.1016/J.CELL.2019.06.029 Publisher Elsevier BV Version Author's final manuscript Citable link https://hdl.handle.net/1721.1/126822 Terms of Use Creative Commons Attribution-NonCommercial-NoDerivs License Detailed Terms http://creativecommons.org/licenses/by-nc-nd/4.0/ HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Cell. Author Manuscript Author manuscript; Manuscript Author available in PMC 2020 July 25. Published in final edited form as: Cell. 2019 July 25; 178(3): 714–730.e22. doi:10.1016/j.cell.2019.06.029. Cellular and inter-cellular rewiring of the human colon during ulcerative colitis Christopher S. Smillie1,19, Moshe Biton1,2,19, José Ordovas-Montañes1,3,4,5,6,7,19, Keri M. Sullivan8, Grace Burgin1, Daniel B. Graham2,8,9,10,11, Rebecca H. Herbst1,12, Noga Rogel1, Michal Slyper1, Julia Waldman1, Malika Sud1, Elizabeth Andrews8, Gabriella Velonias8, Adam L. Haber1, Karthik Jagadeesh1, Sanja Vickovic1, Junmei Yao14, Christine Stevens9, Danielle Dionne1, Lan T. Nguyen1, Alexandra-Chloé Villani1,13, Matan Hofree1, Elizabeth A. Creasey14, Hailiang Huang15,16, Orit Rozenblatt-Rosen1, John J. Garber8, Hamed Khalili8, A. Nicole Desch9,14, Mark J. Daly15,16,17, Ashwin N. Ananthakrishnan8,*, Alex K. Shalek1,3,4,5,6,*, Ramnik J. -

( 12 ) United States Patent ( 10 ) Patent No .: US 10,837,019 B2 Kochenderfer ( 45 ) Date of Patent : * Nov
US010837019B2 ( 12 ) United States Patent ( 10 ) Patent No .: US 10,837,019 B2 Kochenderfer ( 45 ) Date of Patent : * Nov . 17 , 2020 ( 54 ) CHIMERIC ANTIGEN RECEPTORS ( 56 ) References Cited TARGETING B - CELL MATURATION ANTIGEN U.S. PATENT DOCUMENTS 4,235,871 A 11/1980 Papahadjopoulos et al . ( 71 ) Applicant: The United States of America , as 4,501,728 A 2/1985 Geho et al . represented by the Secretary, 4,837,028 A 6/1989 Allen 5,019,369 A 5/1991 Presant et al . Department of Health and Human 5,087,616 A 2/1992 Myers et al . Services, Bethesda, MD (US ) 5,122,464 A 6/1992 Wilson et al . 5,464,758 A 11/1995 Gossen et al . ( 72 ) Inventor: James N. Kochenderfer , Bethesda , 5,770,359 A 6/1998 Wilson et al . 5,814,618 A 9/1998 Bujard et al. MD ( US ) 7,112,715 B2 9/2006 Chambon et al . 9,765,342 B2 9/2017 Kochenderfer ( 73 ) Assignee : The United States of America , as 2007/0009518 A1 1/2007 Novobrantseva et al . represented by the Secretary, 2009/0093024 A1 4/2009 Bowers et al . Department of Health and Human 2011/0020343 Al 1/2011 Senter et al . 2011/0135639 Al 6/2011 Yu et al . Services , Bethesda, MD (US ) 2012/0148552 A1 6/2012 Jensen 2013/0280221 A1 10/2013 Schonfeld et al . ( * ) Notice : Subject to any disclaimer , the term of this 2013/0287748 A1 10/2013 June et al . patent is extended or adjusted under 35 2018/0051292 A1 * 2/2018 Kochenderfer .. CO7K 14/70517 U.S.C. -
GM-CSF Receptor-Beta, Human, Recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor Beta
GM-CSF Receptor-beta, human, recombinant Recombinant Human Granulocyte/Macrophage Colony Stimulating Factor Receptor beta Instruction Manual Catalog Number C-60430 Synonyms CSF2RB,Colony Stimulating Factor 2 Receptor, Beta, Low-Affinity (Granulocyte-Macrophage), GM-CSF/IL-3/IL-5 Receptor Common Beta Subunit, CDw131, IL3RB, SMDP5, IL5RB, Interleukin 3 Receptor/Granulocyte-Macrophage Colony Stimulating Factor 3 Receptor, Beta (High Affinity), Colony-Stimulating Factor-2 Receptor, Beta, Low-Affinity, GM-CSF/IL-3/IL-5 Receptor Common Beta-Chain, Cytokine Receptor Common Subunit Beta, CD131 Antigen, CD131 Description GM-CSF Receptor-beta, also known as CSF2RB is a member of the type I cytokine receptor family. CSF2RB is a high affinity receptor for interleukin-3, interleukin-5 as well as granulocyte- macrophage colony-stimulating factor. The CSF2RB unique form of receptor assembly applies also to IL-3 and IL-5 receptors, providing a structural basis for understanding their activation mechanism which is essential for the development of therapeutics. Human recombinant GM-CSF Receptor-beta produced in Sf9 cells is a single, glycosylated polypeptide chain of 435 amino acids (17-443 a.a), having a molecular mass of 49.7 kDa (migrates at 40-57 kDa on SDS-PAGE under reducing conditions). CSF2RB is fused to an 8 amino acid His-tag at the C-terminus and purified using proprietary chromatographic techniques. Quantity 10 µg Molecular Mass 49.7 kDa Source Sf9 insect cells Biological-Activity NA Specific Activity NA Formulation Sterile-filtered, clear protein solution (0.5 mg/ml) containing phosphate-buffered saline (pH 7.4) and 10% glycerol. Reconstitution Please Note: Always centrifuge product briefly before opening vial. -

How Relevant Are Bone Marrow-Derived Mast Cells (Bmmcs) As Models for Tissue Mast Cells? a Comparative Transcriptome Analysis of Bmmcs and Peritoneal Mast Cells
cells Article How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells 1, 2, 1 1 2,3 Srinivas Akula y , Aida Paivandy y, Zhirong Fu , Michael Thorpe , Gunnar Pejler and Lars Hellman 1,* 1 Department of Cell and Molecular Biology, Uppsala University, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (S.A.); [email protected] (Z.F.); [email protected] (M.T.) 2 Department of Medical Biochemistry and Microbiology, Uppsala University, The Biomedical Center, Box 589, SE-751 23 Uppsala, Sweden; [email protected] (A.P.); [email protected] (G.P.) 3 Department of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Box 7011, SE-75007 Uppsala, Sweden * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 These authors contributed equally to this work. y Received: 29 July 2020; Accepted: 16 September 2020; Published: 17 September 2020 Abstract: Bone marrow-derived mast cells (BMMCs) are often used as a model system for studies of the role of MCs in health and disease. These cells are relatively easy to obtain from total bone marrow cells by culturing under the influence of IL-3 or stem cell factor (SCF). After 3 to 4 weeks in culture, a nearly homogenous cell population of toluidine blue-positive cells are often obtained. However, the question is how relevant equivalents these cells are to normal tissue MCs. By comparing the total transcriptome of purified peritoneal MCs with BMMCs, here we obtained a comparative view of these cells. -
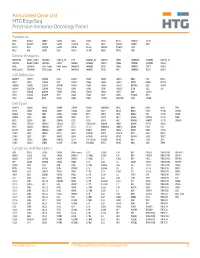
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2 -

The Interleukin 22 Pathway Interacts with Mutant KRAS to Promote Poor Prognosis in Colon Cancer
Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. The interleukin 22 pathway interacts with mutant KRAS to promote poor prognosis in colon cancer Authors: Sarah McCuaig1, David Barras,2, Elizabeth Mann1, Matthias Friedrich1, Samuel Bullers1, Alina Janney1, Lucy C. Garner1, Enric Domingo3, Viktor Hendrik Koelzer3,4,5, Mauro Delorenzi2,6,7, Sabine Tejpar8, Timothy Maughan9, Nathaniel R. West1, Fiona Powrie1 Affiliations: 1 Kennedy Institute of Rheumatology, University of Oxford, Oxford UK. 2 SIB Swiss Institute of Bioinformatics, Bioinformatics Core Facility, Lausanne, Switzerland. 3Department of Oncology, University of Oxford, Oxford UK. 4Nuffield Department of Medicine, University of Oxford, Oxford UK. 5Department of Pathology and Molecular Pathology, University and University Hospital Zurich, Zurich Switzerland. 6 Ludwig Center for Cancer Research, University of Lausanne, Lausanne, Switzerland. 7 Department of Oncology, Faculty of Biology and Medicine, University of Lausanne, Lausanne Switzerland. 8 Molecular Digestive Oncology, KU Leuven, Belgium. 9 CRUK/MRC Oxford Institute for Radiation Oncology, University of Oxford, Oxford, UK. Downloaded from clincancerres.aacrjournals.org on September 26, 2021. © 2020 American Association for Cancer Research. Author Manuscript Published OnlineFirst on May 19, 2020; DOI: 10.1158/1078-0432.CCR-19-1086 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Correspondence to: Professor Fiona Powrie; Kennedy Institute of Rheumatology, University of Oxford, Roosevelt Drive, Headington, Oxford, OX3 7YF, UK. Email: [email protected] Conflicts of Interest: S.M., N.R.W., and F.P. -

The Interleukin-18 Receptor Is Differentially Expressed in Whole
1 The interleukin-18 receptor is differentially expressed in the blood during sepsis. 2 3 Shahan Mamoor, MS1 4 [email protected] East Islip, NY, USA 5 6 We probed published and public microarray datasets1,2 to discover the most significant gene expression changes in the blood of patients with sepsis. We found significant induction 7 expression of IL18RAP and IL18R1, genes encoding subunits of the interleukin-18 receptor, in 8 whole blood from patients with sepsis. 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 Keywords: sepsis, septic shock, interleukin-18, IL18R1, IL18RAP, systems biology of septic 26 shock. 27 28 PAGE 1 OF 13 1 Septic shock is a leading cause of mortality in the United States and worldwide3. We 2 used published and public microarray datasets1,2 to identify differentially expressed genes in the 3 4 blood of patients with sepsis. We identified IL18R1 and IL18RAP as among the genes most 5 differentially expressed in blood in the septic state. 6 7 Methods 8 9 We utilized microarray datasets GSE1001591 and GSE264402 for this differential gene 10 11 expression analysis of blood cells during sepsis. GSE100509 was generated with whole blood 12 using Illumina HumanWG-6 v3.0 expression beadchip technology with n=12 whole blood from 13 control subjects and n=33 whole blood from sepsis patients. GSE26440 was generated using 14 15 Affymetrix Human Genome U133 Plus 2.0 Array technology with n=32 control subjects and 16 n=98 sepsis patients. The Benjamini and Hochberg method of p-value adjustment was used for 17 ranking of differential expression but raw p-values were used for assessment of statistical 18 19 significance of global differential expression. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

Modulation of the IL-33/IL-13 Axis in Obesity by IL-13Rα2
Modulation of the IL-33/IL-13 Axis in Obesity by IL-13R α2 Jennifer Duffen, Melvin Zhang, Katherine Masek-Hammerman, Angela Nunez, Agnes Brennan, Jessica This information is current as E. C. Jones, Jeffrey Morin, Karl Nocka and Marion Kasaian of September 29, 2021. J Immunol published online 5 January 2018 http://www.jimmunol.org/content/early/2018/01/05/jimmun ol.1701256 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2018/01/05/jimmunol.170125 Material 6.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 29, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2018 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published January 5, 2018, doi:10.4049/jimmunol.1701256 The Journal of Immunology Modulation of the IL-33/IL-13 Axis in Obesity by IL-13Ra2 Jennifer Duffen,* Melvin Zhang,* Katherine Masek-Hammerman,† Angela Nunez,‡ Agnes Brennan,* Jessica E. C. Jones,x Jeffrey Morin,‡ Karl Nocka,* and Marion Kasaian* In obesity, IL-13 overcomes insulin resistance by promoting anti-inflammatory macrophage differentiation in adipose tissue. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int.