Gene Expression Profiling of Human Sarcomas: Insights Into Sarcoma Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -
Primepcr™Assay Validation Report
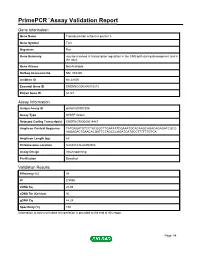
PrimePCR™Assay Validation Report Gene Information Gene Name Transducin-like enhancer protein 3 Gene Symbol Tle3 Organism Rat Gene Summary may be involved in transcription regulation in the CNS both during development and in the adult Gene Aliases Not Available RefSeq Accession No. NM_053400 UniGene ID Rn.24106 Ensembl Gene ID ENSRNOG00000013013 Entrez Gene ID 84424 Assay Information Unique Assay ID qRnoCID0007306 Assay Type SYBR® Green Detected Coding Transcript(s) ENSRNOT00000018467 Amplicon Context Sequence TATGAGATGTCCTACGGCTTGAATATCGAAATGCACAAGCAGACAGAGATCGCG AAGAGACTGAACACGATTCTAGCCCAGATCATGCCTTTTTTGTCA Amplicon Length (bp) 69 Chromosome Location 8:66041636-66059306 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9996 cDNA Cq 20.98 cDNA Tm (Celsius) 80 gDNA Cq 44.24 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Tle3, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Newly Identified Gon4l/Udu-Interacting Proteins
www.nature.com/scientificreports OPEN Newly identifed Gon4l/ Udu‑interacting proteins implicate novel functions Su‑Mei Tsai1, Kuo‑Chang Chu1 & Yun‑Jin Jiang1,2,3,4,5* Mutations of the Gon4l/udu gene in diferent organisms give rise to diverse phenotypes. Although the efects of Gon4l/Udu in transcriptional regulation have been demonstrated, they cannot solely explain the observed characteristics among species. To further understand the function of Gon4l/Udu, we used yeast two‑hybrid (Y2H) screening to identify interacting proteins in zebrafsh and mouse systems, confrmed the interactions by co‑immunoprecipitation assay, and found four novel Gon4l‑interacting proteins: BRCA1 associated protein‑1 (Bap1), DNA methyltransferase 1 (Dnmt1), Tho complex 1 (Thoc1, also known as Tho1 or HPR1), and Cryptochrome circadian regulator 3a (Cry3a). Furthermore, all known Gon4l/Udu‑interacting proteins—as found in this study, in previous reports, and in online resources—were investigated by Phenotype Enrichment Analysis. The most enriched phenotypes identifed include increased embryonic tissue cell apoptosis, embryonic lethality, increased T cell derived lymphoma incidence, decreased cell proliferation, chromosome instability, and abnormal dopamine level, characteristics that largely resemble those observed in reported Gon4l/udu mutant animals. Similar to the expression pattern of udu, those of bap1, dnmt1, thoc1, and cry3a are also found in the brain region and other tissues. Thus, these fndings indicate novel mechanisms of Gon4l/ Udu in regulating CpG methylation, histone expression/modifcation, DNA repair/genomic stability, and RNA binding/processing/export. Gon4l is a nuclear protein conserved among species. Animal models from invertebrates to vertebrates have shown that the protein Gon4-like (Gon4l) is essential for regulating cell proliferation and diferentiation. -

A SARS-Cov-2 Protein Interaction Map Reveals Targets for Drug Repurposing
Article A SARS-CoV-2 protein interaction map reveals targets for drug repurposing https://doi.org/10.1038/s41586-020-2286-9 A list of authors and affiliations appears at the end of the paper Received: 23 March 2020 Accepted: 22 April 2020 A newly described coronavirus named severe acute respiratory syndrome Published online: 30 April 2020 coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19), has infected over 2.3 million people, led to the death of more than Check for updates 160,000 individuals and caused worldwide social and economic disruption1,2. There are no antiviral drugs with proven clinical efcacy for the treatment of COVID-19, nor are there any vaccines that prevent infection with SARS-CoV-2, and eforts to develop drugs and vaccines are hampered by the limited knowledge of the molecular details of how SARS-CoV-2 infects cells. Here we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identifed the human proteins that physically associated with each of the SARS-CoV-2 proteins using afnity-purifcation mass spectrometry, identifying 332 high-confdence protein–protein interactions between SARS-CoV-2 and human proteins. Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (of which, 29 drugs are approved by the US Food and Drug Administration, 12 are in clinical trials and 28 are preclinical compounds). We screened a subset of these in multiple viral assays and found two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the sigma-1 and sigma-2 receptors. -

Loss of TLE3 Promotes the Mitochondrial Program in Beige Adipocytes and Improves Glucose Metabolism
Downloaded from genesdev.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism Stephanie Pearson,1 Anne Loft,2 Prashant Rahbhandari,3 Judith Simcox,1 Sanghoon Lee,1 Peter Tontonoz,3 Susanne Mandrup,2 and Claudio J. Villanueva1 1Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, Utah 84112, USA; 2Department of Biochemistry and Molecular Biology, University of Southern Denmark, 5230 Odense M, Denmark; 3Department of Pathology and Laboratory Medicine, University of California at Los Angeles, Los Angeles, California 90095, USA Prolonged cold exposure stimulates the recruitment of beige adipocytes within white adipose tissue. Beige adipo- cytes depend on mitochondrial oxidative phosphorylation to drive thermogenesis. The transcriptional mechanisms that promote remodeling in adipose tissue during the cold are not well understood. Here we demonstrate that the transcriptional coregulator transducin-like enhancer of split 3 (TLE3) inhibits mitochondrial gene expression in beige adipocytes. Conditional deletion of TLE3 in adipocytes promotes mitochondrial oxidative metabolism and increases energy expenditure, thereby improving glucose control. Using chromatin immunoprecipitation and deep sequencing, we found that TLE3 occupies distal enhancers in proximity to nuclear-encoded mitochondrial genes and that many of these binding sites are also enriched for early B-cell factor (EBF) transcription factors. TLE3 interacts with EBF2 and blocks its ability to promote the thermogenic transcriptional program. Collectively, these studies demonstrate that TLE3 regulates thermogenic gene expression in beige adipocytes through inhibition of EBF2 transcriptional activity. Inhibition of TLE3 may provide a novel therapeutic approach for obesity and diabetes. -

Parental-To-Embryo Switch of Chromosome Organization in Early
Parental-to-embryo switch of chromosome organization in early embryogenesis Samuel Collombet, Noemie Ranisavljevic, Takashi Nagano, Csilla Varnai, Tarak Shisode, Wing Leung, Tristan Piolot, Rafael Galupa, Maud Borensztein, Nicolas Servant, et al. To cite this version: Samuel Collombet, Noemie Ranisavljevic, Takashi Nagano, Csilla Varnai, Tarak Shisode, et al.. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature, Nature Pub- lishing Group, 2020, 580, pp.142 - 146. 10.1038/s41586-020-2125-z. hal-03027197 HAL Id: hal-03027197 https://hal.archives-ouvertes.fr/hal-03027197 Submitted on 27 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Article Parental-to-embryo switch of chromosome organization in early embryogenesis https://doi.org/10.1038/s41586-020-2125-z Samuel Collombet1,2,10, Noémie Ranisavljevic1,3,10, Takashi Nagano4,9,10, Csilla Varnai4,5, Tarak Shisode6, Wing Leung4,9, Tristan Piolot1, Rafael Galupa1,2, Maud Borensztein1, Received: 3 April 2019 Nicolas Servant7, Peter Fraser4,8,11 ✉, Katia Ancelin1,11 ✉ & Edith Heard1,2,11 ✉ Accepted: 16 January 2020 Published online: 25 March 2020 Paternal and maternal epigenomes undergo marked changes after fertilization1. Recent Check for updates epigenomic studies have revealed the unusual chromatin landscapes that are present in oocytes, sperm and early preimplantation embryos, including atypical patterns of histone modifcations2–4 and diferences in chromosome organization and accessibility, both in gametes5–8 and after fertilization5,8–10. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Identification of Differentially Methylated Region (DMR) Networks Associated with Progression of Nonalcoholic Fatty Liver Disease
www.nature.com/scientificreports OPEN Identifcation of diferentially methylated region (DMR) networks associated with progression of Received: 16 March 2018 Accepted: 29 August 2018 nonalcoholic fatty liver disease Published: xx xx xxxx Kikuko Hotta 1, Aya Kitamoto2, Takuya Kitamoto2, Yuji Ogawa3, Yasushi Honda3, Takaomi Kessoku3, Masato Yoneda3, Kento Imajo3, Wataru Tomeno3,4, Satoru Saito3 & Atsushi Nakajima3 The progression of nonalcoholic fatty liver disease (NAFLD) is afected by epigenetics. We performed diferentially methylated region (DMR) and co-methylation analyses to identify DMR networks associated with the progression of NAFLD. DMRs displaying diferences in multiple consecutive diferentially methylated CpGs between mild and advanced NAFLD were extracted. The average values of topological overlap measures for the CpG matrix combining two diferent DMRs were calculated and two DMR networks that strongly correlated with the stages of fbrosis were identifed. The annotated genes of one network included genes involved in transcriptional regulation, cytoskeleton organization, and cellular proliferation. The annotated genes of the second network were primarily associated with metabolic pathways. The CpG methylation levels in these networks were strongly afected by age and fasting plasma glucose levels, which may be important co-regulatory factors. The methylation status of fve DMRs in the second network was reversible following weight loss. Our results suggest that CpG methylation in DMR networks is regulated concomitantly via aging -

Meta-Analysis of Human Cancer Microarrays Reveals GATA3 Is Integral to the Estrogen Receptor Alpha Pathway Brian J Wilson and Vincent Giguère*
Molecular Cancer BioMed Central Research Open Access Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway Brian J Wilson and Vincent Giguère* Address: Molecular Oncology Group, Room H5-45, McGill University Health Centre, 687 Pine Avenue West, Montréal, Québec, H3A 1A1, Canada Email: Brian J Wilson - [email protected]; Vincent Giguère* - [email protected] * Corresponding author Published: 4 June 2008 Received: 28 November 2007 Accepted: 4 June 2008 Molecular Cancer 2008, 7:49 doi:10.1186/1476-4598-7-49 This article is available from: http://www.molecular-cancer.com/content/7/1/49 © 2008 Wilson and Giguère; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: The transcription factor GATA3 has recently been shown to be necessary for mammary gland morphogenesis and luminal cell differentiation. There is also an increasing body of data linking GATA3 to the estrogen receptor α (ERα) pathway. Among these it was shown that GATA3 associates with the promoter of the ERα gene and ERα can reciprocally associate with the GATA3 gene. GATA3 has also been directly implicated in a differentiated phenotype in mouse models of mammary tumourigenesis. The purpose of our study was to compare coexpressed genes, by meta-analysis, of GATA3 and relate these to a similar analysis for ERα to determine the depth of overlap. Results: We have used a newly described method of meta-analysis of multiple cancer studies within the Oncomine database, focusing here predominantly upon breast cancer studies. -

Analysis of Key Genes and Pathways Associated with the Pathogenesis of Type 2 Diabetes Mellitus
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.12.456106; this version posted August 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Analysis of key genes and pathways associated with the pathogenesis of Type 2 diabetes mellitus Varun Alur1, Varshita Raju2, Basavaraj Vastrad3, Chanabasayya Vastrad*4, Shivakumar Kotturshetti4 1. Department of Endocrinology, J.J. M Medical College, Davanagere, Karnataka 577004, India. 2. Department of Obstetrics and Gynecology, J.J. M Medical College, Davanagere, Karnataka 577004, India. 3. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 4. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.08.12.456106; this version posted August 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Type 2 diabetes mellitus (T2DM) is the most common endocrine disorder which poses a serious threat to human health. This investigation aimed to screen the candidate genes differentially expressed in T2DM by bioinformatics analysis. The expression profiling by high throughput sequencing of GSE81608 dataset was retrieved from the gene expression omnibus (GEO) database and analyzed to identify the differentially expressed genes (DEGs) between T2DM and normal controls. Then, Gene Ontology (GO) and pathway enrichment analysis, protein- protein interaction (PPI) network, modules, miRNA-hub gene regulatory network construction and TF-hub gene regulatory network construction, and topological analysis were performed. -

Viruses 2015, 7, 2428-2449; Doi:10.3390/V7052428 OPEN ACCESS
Viruses 2015, 7, 2428-2449; doi:10.3390/v7052428 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Article Impact of Adenovirus E4-ORF3 Oligomerization and Protein Localization on Cellular Gene Expression Elizabeth I. Vink 1,†, Yueting Zheng 1, Rukhsana Yeasmin 2, Thomas Stamminger 3, Laurie T. Krug 1 and Patrick Hearing 1,* 1 Department of Molecular Genetics and Microbiology, Stony Brook University, Stony Brook, NY 11794-5222, USA; E-Mails: [email protected] (Y.Z.); [email protected] (L.T.K.) 2 Department of Computer Science, Stony Brook University, Stony Brook, NY 11794, USA; E-Mail: [email protected] 3 Institute for Clinical and Molecular Virology, University of Erlangen-Nuremberg, Schlossgarten 4, Erlangen 91054, Germany; E-Mail: [email protected] † Current address: Department of Microbiology, New York University Langone Medical Center, New York, NY 10016, USA; E-Mail: [email protected]. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-631-632-8813; Fax: +1-631-632-9797. Academic Editor: Joanna Parish Received: 31 March 2015 / Accepted: 11 May 2015 / Published: 13 May 2015 Abstract: The Adenovirus E4-ORF3 protein facilitates virus replication through the relocalization of cellular proteins into nuclear inclusions termed tracks. This sequestration event disrupts antiviral properties associated with target proteins. Relocalization of Mre11-Rad50-Nbs1 proteins prevents the DNA damage response from inhibiting Ad replication. Relocalization of PML and Daxx impedes the interferon-mediated antiviral response. Several E4-ORF3 targets regulate gene expression, linking E4-ORF3 to transcriptional control. -
Expression Profiling of WD40 Family Genes Including DDB1- and CUL4
Mistry et al. BMC Genomics (2020) 21:602 https://doi.org/10.1186/s12864-020-07016-9 RESEARCH ARTICLE Open Access Expression profiling of WD40 family genes including DDB1- and CUL4- associated factor (DCAF) genes in mice and human suggests important regulatory roles in testicular development and spermatogenesis Bhavesh V. Mistry1, Maha Alanazi1, Hanae Fitwi1,2, Olfat Al-Harazi3, Mohamed Rajab1, Abdullah Altorbag1, Falah Almohanna1, Dilek Colak2 and Abdullah M. Assiri1,3,4* Abstract Background: The WD40-repeat containing proteins, including DDB1–CUL4-associated factors (DCAFs), are abundant and conserved proteins that play important roles in different cellular processes including spermatogenesis. DCAFs are subset of WD40 family proteins that contain WDxR motif and have been proposed to function as substrate receptor for Cullin4-RING-based E3 ubiquitin ligase complexes to recruit diverse proteins for ubiquitination, a vital process in spermatogenesis. Large number of WD40 genes has been identified in different species including mouse and human. However, a systematic expression profiling of WD40 genes in different tissues of mouse and human has not been investigated. We hypothesize that large number of WD40 genes may express highly or specifically in the testis, where their expression is uniquely regulated during testis development and spermatogenesis. Therefore, the objective of this study is to mine and characterize expression patterns of WD40 genes in different tissues of mouse and human with particular emphasis on DCAF genes expressions during mouse testicular development. (Continued on next page) * Correspondence: [email protected] 1Department of Comparative Medicine, King Faisal Specialist Hospital & Research Centre, Riyadh, Saudi Arabia 3Biostatistics, Epidemiology and Scientific Computing Department, King Faisal Specialist Hospital & Research Centre, Riyadh, Saudi Arabia Full list of author information is available at the end of the article © The Author(s).