Loss of TLE3 Promotes the Mitochondrial Program in Beige Adipocytes and Improves Glucose Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

I LITERATURE-BASED DISCOVERY of KNOWN and POTENTIAL NEW
LITERATURE-BASED DISCOVERY OF KNOWN AND POTENTIAL NEW MECHANISMS FOR RELATING THE STATUS OF CHOLESTEROL TO THE PROGRESSION OF BREAST CANCER BY YU WANG THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics with a concentration in Library and Information Science in the Graduate College of the University of Illinois at Urbana-Champaign, 2019 Urbana, Illinois Adviser: Professor Vetle I. Torvik Professor Erik Russell Nelson i ABSTRACT Breast cancer has been studied for a long period of time and from a variety of perspectives in order to understand its pathogeny. The pathogeny of breast cancer can be classified into two groups: hereditary and spontaneous. Although cancer in general is considered a genetic disease, spontaneous factors are responsible for most of the pathogeny of breast cancer. In other words, breast cancer is more likely to be caused and deteriorated by the dysfunction of a physical molecule than be caused by germline mutation directly. Interestingly, cholesterol, as one of those molecules, has been discovered to correlate with breast cancer risk. However, the mechanisms of how cholesterol helps breast cancer progression are not thoroughly understood. As a result, this study aims to study known and discover potential new mechanisms regarding to the correlation of cholesterol and breast cancer progression using literature review and literature-based discovery. The known mechanisms are further classified into four groups: cholesterol membrane content, transport of cholesterol, cholesterol metabolites, and other. The potential mechanisms, which are intended to provide potential new treatments, have been identified and checked for feasibility by an expert. -

Uncoupling Proteins: Functional Characteristics and Role in the Pathogenesis of Obesity and Type II Diabetes
Diabetologia 2001) 44: 946±965 Ó Springer-Verlag 2001 Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and Type II diabetes L. T.Dalgaard, O.Pedersen Steno Diabetes Center, Gentofte, Denmark Abstract obesity. The three uncoupling protein homologue genes UCP1, UCP2, and UCP3 have been investigat- Uncoupling proteins are mitochondrial carrier pro- ed for polymorphisms and mutations and their impact teins which are able to dissipate the proton gradient on Type II diabetes mellitus, obesity, and body weight of the inner mitochondrial membrane. This uncou- gain or BMI. The main conclusion is that variation in pling process reduces the amount of ATP generated the UCP1, UCP2 or UCP3 genes is not associated through an oxidation of fuels. The hypothesis that un- with major alterations of body weight gain. The con- coupling proteins UCPs) are candidate genes for hu- tribution of UCP genes towards polygenic obesity man obesity or Type II non-insulin-dependent) dia- and Type II diabetes is evaluated and discussed. [Dia- betes mellitus is based on the finding that a chemical betologia 2001) 44: 946±965] uncoupling of the mitochondrial membrane reduces body adiposity, and that lower metabolic rates predict Keywords Uncoupling proteins, Type II diabetes weight gain. It is straightforward to hypothesize that mellitus, obesity, genetics, body weight regulation, common polymorphisms of UCP1, UCP2 and UCP3 energy expenditure, metabolic rate, brown adipose genes lower metabolic rate by a more efficient energy tissue, white adipose tissue, reactive oxygen species, coupling in the mitochondria. Furthermore, geneti- polymorphism, mutation, transgenics, gene knock- cally engineered mice over expressing different UCP out. -

Nrg4 and Gpr120 Signalling in Brown Fat Anthony Chukunweike OKOLO
Nrg4 and Gpr120 Signalling in Brown Fat Anthony Chukunweike OKOLO Institute of Reproductive and Developmental Biology Department of Surgery and Cancer Faculty of Medicine Imperial College London A thesis submitted in fulfilment of the requirements for award of the degree of Doctor of Philosophy 1 Statement of Originality All experiments included in this thesis were performed by me unless otherwise stated in the text. 2 Copyright statement The copyright of this thesis rests with the author and is made available under a Creative Commons Attribution Non-Commercial No Directive Licence. Researchers are free to copy, distribute or transmit the thesis on the condition that they attribute it, and they do not use it for commercial purposes, and they do not alter, transform or build upon it. For any re-use or re-distribution, researchers must make clear to others the licence terms of this work. 3 Acknowledgments I would like to thank my supervisors Dr Aylin Hanyaloglu and Dr Mark Christian for giving me the great opportunity to work in their labs. Aylin put in a great deal of effort especially in area of Gpr120 signalling, including having to guide me through the critical imaging procedures. Aylin and Mark contributed a great deal towards the final edition of this thesis. I would also like to thank Dr Mark Christian for bringing me to Imperial College London to start off my PhD in his laboratory, and for being a great mentor and a continuous source of knowledge for me. I am grateful for your enduring patience in trying to bring out the best in me and ensuring that I develop the ‘critical thinking’ that is needed as a scientist. -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -
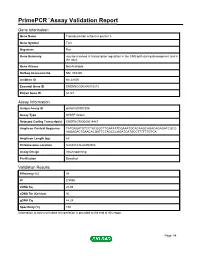
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name Transducin-like enhancer protein 3 Gene Symbol Tle3 Organism Rat Gene Summary may be involved in transcription regulation in the CNS both during development and in the adult Gene Aliases Not Available RefSeq Accession No. NM_053400 UniGene ID Rn.24106 Ensembl Gene ID ENSRNOG00000013013 Entrez Gene ID 84424 Assay Information Unique Assay ID qRnoCID0007306 Assay Type SYBR® Green Detected Coding Transcript(s) ENSRNOT00000018467 Amplicon Context Sequence TATGAGATGTCCTACGGCTTGAATATCGAAATGCACAAGCAGACAGAGATCGCG AAGAGACTGAACACGATTCTAGCCCAGATCATGCCTTTTTTGTCA Amplicon Length (bp) 69 Chromosome Location 8:66041636-66059306 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9996 cDNA Cq 20.98 cDNA Tm (Celsius) 80 gDNA Cq 44.24 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Tle3, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Perspectives in Diabetes Uncoupling Proteins 2 and 3 Potential Regulators of Mitochondrial Energy Metabolism Olivier Boss, Thilo Hagen, and Bradford B
Perspectives in Diabetes Uncoupling Proteins 2 and 3 Potential Regulators of Mitochondrial Energy Metabolism Olivier Boss, Thilo Hagen, and Bradford B. Lowell Mitochondria use energy derived from fuel combustion fuels and oxygen are converted into carbon dioxide, water, to create a proton electrochemical gradient across the and ATP (Fig. 1). The key challenge for the organism is to reg- mitochondrial inner membrane. This intermediate form ulate these many steps so that rates of ATP production are of energy is then used by ATP synthase to synthesize equal to rates of ATP utilization. This is not a small task given AT P. Uncoupling protein-1 (UCP1) is a brown fat–spe- that rates of ATP utilization can quickly increase severalfold cific mitochondrial inner membrane protein with proton (up to 100-fold in muscle during contraction). transport activity. UCP1 catalyzes a highly regulated proton leak, converting energy stored within the mito- Fuel metabolism and oxidative phosphorylation consist chondrial proton electrochemical potential gradient to of many tightly coupled enzymatic reactions (Fig. 1), which heat. This uncouples fuel oxidation from conversion of are regulated, in part, by ADP availability. Control by ADP is ADP to AT P. In rodents, UCP1 activity and brown fat accounted for by the chemiosmotic hypothesis of Mitchell (1). contribute importantly to whole-body energy expendi- Oxidation of fuels via the electron transport chain generates ture. Recently, two additional mitochondrial carriers a proton electrochemical potential gradient ( µH+) across with high similarity to UCP1 were molecularly cloned. the mitochondrial inner membrane. Protons reenter the In contrast to UCP1, UCP2 is expressed widely, and mitochondrial matrix via ATP synthase (F0F1-A TPase) in a UCP3 is expressed preferentially in skeletal muscle. -

Gene Expression Profiling of Human Sarcomas: Insights Into Sarcoma Biology
Research Article Gene Expression Profiling of Human Sarcomas: Insights into Sarcoma Biology Kristin Baird,1 Sean Davis,1 Cristina R. Antonescu,3 Ursula L. Harper,1 Robert L. Walker,1 Yidong Chen,1 Arthur A. Glatfelter,1 Paul H. Duray,2 and Paul S. Meltzer1 1Cancer Genetics Branch, National Human Genome Research Institute; 2Laboratory of Pathology, National Cancer Institute, NIH, Bethesda, Maryland; 3Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York Abstract changes. This latter group occurs more frequently in older adults and includes several types of sarcomas that lack known disease- Sarcomas are a biologically complex group of tumors of specific chromosome translocations or mutations, but may contain mesenchymal origin. By using gene expression microarray mutations in RB1, CDKN2A, and TP53. Investigation of these and analysis, we aimed to find clues into the cellular differenti- other aspects of sarcoma biology have provided insights into ation and oncogenic pathways active in these tumors as well broadly relevant fundamental mechanisms of oncogenesis (for as potential biomarkers and therapeutic targets. We exam- review, see ref. 1) and may have profound implications in the ined 181 tumors representing 16 classes of human bone and development of therapeutic intervention. This is exemplified by the soft tissue sarcomas on a 12,601-feature cDNA microarray. successful treatment of gastrointestinal stromal tumors, which Remarkably, 2,766 probes differentially expressed across this frequently have activating mutations of KIT, with the tyrosine sample set clearly delineated the various tumor classes. kinase inhibitor, imatinib mesylate. Although genetic alterations, Several genes of potential biological and therapeutic interest particularly fusion genes arising from translocations, have been were associated with each sarcoma type, including specific identified in many sarcomas, the function of the fusion gene tyrosine kinases, transcription factors, and homeobox genes. -

FARE2021WINNERS Sorted by Institute
FARE2021WINNERS Sorted By Institute Swati Shah Postdoctoral Fellow CC Radiology/Imaging/PET and Neuroimaging Characterization of CNS involvement in Ebola-Infected Macaques using Magnetic Resonance Imaging, 18F-FDG PET and Immunohistology The Ebola (EBOV) virus outbreak in Western Africa resulted in residual neurologic abnormalities in survivors. Many case studies detected EBOV in the CSF, suggesting that the neurologic sequelae in survivors is related to viral presence. In the periphery, EBOV infects endothelial cells and triggers a “cytokine stormâ€. However, it is unclear whether a similar process occurs in the brain, with secondary neuroinflammation, neuronal loss and blood-brain barrier (BBB) compromise, eventually leading to lasting neurological damage. We have used in vivo imaging and post-necropsy immunostaining to elucidate the CNS pathophysiology in Rhesus macaques infected with EBOV (Makona). Whole brain MRI with T1 relaxometry (pre- and post-contrast) and FDG-PET were performed to monitor the progression of disease in two cohorts of EBOV infected macaques from baseline to terminal endpoint (day 5-6). Post-necropsy, multiplex fluorescence immunohistochemical (MF-IHC) staining for various cellular markers in the thalamus and brainstem was performed. Serial blood and CSF samples were collected to assess disease progression. The linear mixed effect model was used for statistical analysis. Post-infection, we first detected EBOV in the serum (day 3) and CSF (day 4) with dramatic increases until euthanasia. The standard uptake values of FDG-PET relative to whole brain uptake (SUVr) in the midbrain, pons, and thalamus increased significantly over time (p<0.01) and positively correlated with blood viremia (p≤0.01). -

Newly Identified Gon4l/Udu-Interacting Proteins
www.nature.com/scientificreports OPEN Newly identifed Gon4l/ Udu‑interacting proteins implicate novel functions Su‑Mei Tsai1, Kuo‑Chang Chu1 & Yun‑Jin Jiang1,2,3,4,5* Mutations of the Gon4l/udu gene in diferent organisms give rise to diverse phenotypes. Although the efects of Gon4l/Udu in transcriptional regulation have been demonstrated, they cannot solely explain the observed characteristics among species. To further understand the function of Gon4l/Udu, we used yeast two‑hybrid (Y2H) screening to identify interacting proteins in zebrafsh and mouse systems, confrmed the interactions by co‑immunoprecipitation assay, and found four novel Gon4l‑interacting proteins: BRCA1 associated protein‑1 (Bap1), DNA methyltransferase 1 (Dnmt1), Tho complex 1 (Thoc1, also known as Tho1 or HPR1), and Cryptochrome circadian regulator 3a (Cry3a). Furthermore, all known Gon4l/Udu‑interacting proteins—as found in this study, in previous reports, and in online resources—were investigated by Phenotype Enrichment Analysis. The most enriched phenotypes identifed include increased embryonic tissue cell apoptosis, embryonic lethality, increased T cell derived lymphoma incidence, decreased cell proliferation, chromosome instability, and abnormal dopamine level, characteristics that largely resemble those observed in reported Gon4l/udu mutant animals. Similar to the expression pattern of udu, those of bap1, dnmt1, thoc1, and cry3a are also found in the brain region and other tissues. Thus, these fndings indicate novel mechanisms of Gon4l/ Udu in regulating CpG methylation, histone expression/modifcation, DNA repair/genomic stability, and RNA binding/processing/export. Gon4l is a nuclear protein conserved among species. Animal models from invertebrates to vertebrates have shown that the protein Gon4-like (Gon4l) is essential for regulating cell proliferation and diferentiation. -

Irisin As a Multifunctional Protein: Implications for Health and Certain Diseases
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Jagiellonian Univeristy Repository medicina Review Irisin as a Multifunctional Protein: Implications for Health and Certain Diseases Paulina Korta 1 , Ewa Poche´c 1,* and Agnieszka Mazur-Biały 2 1 Department of Glycoconjugate Biochemistry, Institute of Zoology and Biomedical Research, Faculty of Biology, Jagiellonian University, Gronostajowa 9, 30-387 Krakow, Poland 2 Department of Ergonomics and Exercise Physiology, Faculty of Health Sciences, Jagiellonian University, Medical College, Grzegorzecka 20, 31-531 Krakow, Poland * Correspondence: [email protected]; Tel.: +48-12-664-64-67 Received: 29 June 2019; Accepted: 12 August 2019; Published: 15 August 2019 Abstract: Sedentary life style is considered to be an independent risk factor for many disorders, including development of type 2 diabetes, obesity, immune dysfunction, asthma, and neurological or coronary heart disease. Irisin is released from myocytes during physical activity, and acts as a link between muscles and other tissues and organs. This myokine is produced as a result of proteolytic cleavage of FNDC5 protein present in the membrane of myocytes. Secretion of irisin is regulated by N-linked oligosaccharides attached to the protein molecule. The two N-glycan molecules, which constitute a significant part of the irisin glycoprotein, regulate the browning of adipocytes, which is the most important function of irisin. A receptor specific for irisin has still not been discovered. In some tissues irisin probably acts via integrins, which are widely expressed transmembrane receptors. Many studies have confirmed the multifunctional role of irisin and the beneficial effects of this molecule on body homeostasis. -

Exclusive Occurrence of Thermogenin Antigen in Brown Adipose Tissue
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Volume 150, number 1 FEBS LETTERS December 1982 Exclusive occurrence of thermogenin antigen in brown adipose tissue Barbara Cannon, Anders Hedin* and Jan Nedergaard Departments of Metabolic Research and *Immunology, The Wenner-Gren Institute, University of Stockholm, Norrtullsgatan 16, 113 45 Stockholm, Sweden Received 25 October 1982 Thermogenin is the purine-nucleotide binding polypeptide in brown adipose tissue mitochondria (Mr 32000) which confers upon these mitochondria the ability to produce heat. An enzyme-linked immunosorbent assay (ELISA) has been developed to demonstrate and quantitate the occurrence of thermogenin antigen in small amounts of tissue, and thus to characterize different depots of fat tissue as white or brown. The extreme sensitivity of the method allows determination of thermogenin in samples equivalent to < 1 mg tissue. The results indicate that thermogenin seems to be exclusively localised in brown fat mitochondria (as compared to white fat, liver or heart muscle mitochondria), and thermogenin antigen could only be found in brown adipocytes (as compared to white adipocytes). Thus, brown and white adipose tissue are probably ontogenetically different Brown adipose tissue ELISA Mitochondria Nonshivering thermogenesis Thermogenin White adipose tissue 1. INTRODUCTION form of white adipose tissue, or whether it should be considered as a bona fide organ in itself, with its The demonstration that the thermogenic func- own ontogeny [lo]. tion of brown adipose tissue is directly related to The present interest in the connection between the presence in the mitochondria of a specific poly- the activity of brown adipose tissue and the ten- peptide - thermogenin - with a subunit A4, 32000 dency to evolve obesity in animals and man (review has made it possible to study thermogenesis at the [l 11) has made it necessary to be able to charac- molecular level [1,2], (reviews [3-51).