Inhibition of C-Jun N-Terminal Kinase by SP600125: a Cdna Microarray Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Influencers on Thyroid Cancer Onset: Molecular Genetic Basis
G C A T T A C G G C A T genes Review Influencers on Thyroid Cancer Onset: Molecular Genetic Basis Berta Luzón-Toro 1,2, Raquel María Fernández 1,2, Leticia Villalba-Benito 1,2, Ana Torroglosa 1,2, Guillermo Antiñolo 1,2 and Salud Borrego 1,2,* 1 Department of Maternofetal Medicine, Genetics and Reproduction, Institute of Biomedicine of Seville (IBIS), University Hospital Virgen del Rocío/CSIC/University of Seville, 41013 Seville, Spain; [email protected] (B.L.-T.); [email protected] (R.M.F.); [email protected] (L.V.-B.); [email protected] (A.T.); [email protected] (G.A.) 2 Centre for Biomedical Network Research on Rare Diseases (CIBERER), 41013 Seville, Spain * Correspondence: [email protected]; Tel.: +34-955-012641 Received: 3 September 2019; Accepted: 6 November 2019; Published: 8 November 2019 Abstract: Thyroid cancer, a cancerous tumor or growth located within the thyroid gland, is the most common endocrine cancer. It is one of the few cancers whereby incidence rates have increased in recent years. It occurs in all age groups, from children through to seniors. Most studies are focused on dissecting its genetic basis, since our current knowledge of the genetic background of the different forms of thyroid cancer is far from complete, which poses a challenge for diagnosis and prognosis of the disease. In this review, we describe prevailing advances and update our understanding of the molecular genetics of thyroid cancer, focusing on the main genes related with the pathology, including the different noncoding RNAs associated with the disease. -

The Role of the S6K2 Splice Isoform in Mtor/S6K Signalling and Cellular Functions
The role of the S6K2 splice isoform in mTOR/S6K signalling and cellular functions Olena Myronova A thesis submitted to the University College London in fulfilment with the requirements for the degree of Doctor of Philosophy London, November 2015 Research Department of Structural and Molecular Biology Division of Biosciences University College London Gower Street London, WC1E 6BT United Kingdom Ludwig Institute for Cancer Research 666 Third Avenue, 28th floor New York, N.Y. 10017 USA The role of the S6K2 splice isoform in mTOR/S6K signalling and cellular functions 1 Declaration I, Olena Myronova, declare that all the work presented in this thesis is the result of my own work. The work presented here does not constitute part of any other thesis. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. The work here in was carried out while I was a graduate research student at University College London, Research Department of Structural and Molecular Biology under the supervision of Professor Ivan Gout. Olena Myronova The role of the S6K2 splice isoform in mTOR/S6K signalling and cellular functions 2 Abstract Ribosomal S6 kinase (S6K) is a member of the AGC family of serine/threonine protein kinases and plays a key role in diverse cellular processes, including cell growth, survival and metabolism. Activation of S6K by growth factors, amino acids, energy levels and hypoxia is mediated by the mTOR and PI3K signalling pathways. Dysregulation of S6K activity has been implicated in a number of human pathologies, including cancer, diabetes, obesity and ageing. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Xo GENE PANEL
xO GENE PANEL Targeted panel of 1714 genes | Tumor DNA Coverage: 500x | RNA reads: 50 million Onco-seq panel includes clinically relevant genes and a wide array of biologically relevant genes Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 -

Comprehensive Assessment of Indian Variations in the Druggable Kinome Landscape Highlights Distinct Insights at the Sequence, Structure and Pharmacogenomic Stratum
SUPPLEMENTARY MATERIAL Comprehensive assessment of Indian variations in the druggable kinome landscape highlights distinct insights at the sequence, structure and pharmacogenomic stratum Gayatri Panda1‡, Neha Mishra1‡, Disha Sharma2,3, Rahul C. Bhoyar3, Abhinav Jain2,3, Mohamed Imran2,3, Vigneshwar Senthilvel2,3, Mohit Kumar Divakar2,3, Anushree Mishra3, Priyanka Banerjee4, Sridhar Sivasubbu2,3, Vinod Scaria2,3, Arjun Ray1* 1 Department of Computational Biology, Indraprastha Institute of Information Technology, Okhla, India. 2 Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India. 3 CSIR-Institute of Genomics and Integrative Biology, Mathura Road, Delhi-110020, India. 4 Institute for Physiology, Charite-University of Medicine, Berlin, 10115 Berlin, Germany. ‡These authors contributed equally to this work. * [email protected] TABLE OF CONTENTS Name Title Supplemental_Figure_S1 Fauchere and Pliska hydrophobicity scale for variations in structure data Supplemental_Figure_S2 Phenotypic drug-drug correlogram Supplemental_Table_S1 545 kinase coding genes used in the study Supplemental_Table_S2 Classes and count of kinase coding genes Supplemental_Table_S3 Allele frequency Indian v/s other populations from 1000 genome data(1000g2015). Supplemental_Table_S4 IndiGen Structure Data- consisting of 12 genes and their 22 variants Supplemental_Table_S5 Genes, PDB ids, mutations in IndiGen data and associated drugs (FDA approved) Supplemental_Table_S6 Data used for docking and binding pocket similarity analysis Supplemental_Table_S7 -

Molecular Mechanisms Underlying Innate Immune Kinase
MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION APPROVED BY SUPERVISORY COMMITTEE Michael A White, Ph.D. Melanie H. Cobb, Ph.D. Lawrence Lum, Ph.D. John D. Minna, M.D. DEDICATION This work is dedicated to my mother and Arlene for their love and support. ACKNOWLEDGEMENTS I am very grateful to my mentor, Dr. Michael White, for his continuous support and guidance through the entire study. I really appreciate his inspiration, patience, and generosity. I would also like to thank my committee members, Dr. Cobb, Dr. Lum, and Dr. Minna, for their invaluable advice and discussion. I thank all the White lab members and my friends for their help, suggestion, and discussion. Particularly, I would like to thank Rosie, Michael, Brian, Tzuling, and Malia for their long-term support and collaboration. I would also like to thank my friends, Veleka, Pei-Ling, Jen-Chieh, Shu-Yi, Chih-Chiang, Jen-Shuan, Yi-Chun, and Yu-San for their friendship. I am grateful to Drs. Rolf Brekken, Zhijian James Chen, Xuetao Cao, Philip Tsichlis, Charles Yeaman, William Hahn, Keqiang Ye, and Shu-Chan Hsu, Bing Su, Dos Sarbassov, Mark Magnuson, David Sabatini, Thomas Tan, and Bert Vogelstein for many of the reagents used in these studies. Finally, and most importantly, I would like to thank my mother and Arlene for their unending support and encouragement. MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION by YI-HUNG OU DISSERTATION Presented to the Faculty of the Graduate School of Biomedical Sciences The University of Texas Southwestern Medical Center at Dallas In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY The University of Texas Southwestern Medical Center at Dallas Dallas, Texas April, 2013 Copyright by YI-HUNG OU, 2013 All Rights Reserved MOLECULAR MECHANISMS UNDERLYING INNATE IMMUNE KINASE TBK1-DRIVEN ONCOGENIC TRANSFORMATION Publication No. -
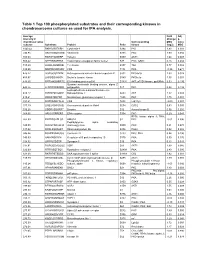
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Members of the Competence Network for Congenital Heart Defects, Germany
Members of the Competence Network for Congenital Heart Defects, Germany Hashim Abdul-Khaliq, Hans-Heiner Kramer, Felix Berger, Brigitte Stiller, Ulrike Bauer, Thomas Pickardt, Sabine Klaassen Family 49 Family 62 Family 226 Family 333 * * * * * * * * TOF AVS ASD ASD PAPVR TOF * * * * ASD PAPVD COA ASD * Family 346 Family 398 VSD * Family 489 * * VSD VSD * * VSD HCM * * * Family 545 BAV AVS ASD PDA Sv AS VSD Family 576 Family 645 Septal defect * * * COA AVS ASD BAV VSD * * * * BAV AVS AVS ASD BAV Family 702 * VSD VSD Family 732 * Family 720 AVSD * * * VSD * * Family 831 TOF TOF TOF Vring TGA VSD * * * * PDA VSD AVS * PDA * * * VSD BAV PFO ASD ASD2 PDA PDA COA Family 1117 Family 1121 * PVS ? * ASD VSD * * * TOF VSD ASD VSD Family 1319 Family 1151 * * * * 2 * ASD ASD * * ASD ASD * * EbA AVS AVS Family 1364 Family 1560 Family 1575 * * TOF VSD ASD VSD infPS TOF * * * VSD PVS PVS PVS VSD * VSD PVS Family 1710 ASD Family 1722 * ASD CHD * VSD VSD * COA TGA DCM ASD BAV SV, MVA COA PVS * DCM DCM ASD HLHS COA BAV Family 2077 Family 2261 * COA, BAV * * * PVS PVS PVS * * infPS infPS BAV ASD * * AVS ASD ASD BAV BAV Family 3500 Family 2558 Family 3315 * * * AVS VSD AVR * * HLHS COA PDA ASD BAV * PVA Family 3501 VSD Family 3503 ? ? * * * VSD HRHS EbA * 3 PVA PAA ASD PDA VSD Family 3505 Family 3540 * * BAV ASD AVS * * * HLHS HLHS TAPVR BAV COA Figure S1. Pedigrees of 32 Danish multiplex CHD families. Circles: females. Squares: males. White symbols: unaffected family members. Filled symbols: affected family members. Triangles: abortion. -

NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-7-2018 Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer Jairo Ramos [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Clinical Epidemiology Commons Recommended Citation Ramos, Jairo, "Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer" (2018). FIU Electronic Theses and Dissertations. 3872. https://digitalcommons.fiu.edu/etd/3872 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida DECIPHER MECHANISMS BY WHICH NUCLEAR RESPIRATORY FACTOR ONE (NRF1) COORDINATES CHANGES IN THE TRANSCRIPTIONAL AND CHROMATIN LANDSCAPE AFFECTING DEVELOPMENT AND PROGRESSION OF INVASIVE BREAST CANCER A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in PUBLIC HEALTH by Jairo Ramos 2018 To: Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work This dissertation, Written by Jairo Ramos, and entitled Decipher Mechanisms by Which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer, having been approved in respect to style and intellectual content, is referred to you for judgment. -

Supplemental File
SUPPLEMENTARY MATERIALS AND METHODS ### reorder as minimun to maximum relative induction. allks2=cbind(allks,apply(allks,1,sum)) For the different analyses, the macros are as following: allks3=allks2[order(allks2[,length(colnames(allks2))]),] allks5=allks3[,-length(colnames(allks2))] -Heatmap and correlation analyses # check allks5 source("http://bioconductor.org/biocLite.R") biocLite() -Heatmap install.packages("gplots") library("gplots") #Set a color scale "zero centered" #Set Graph title (do one by one) breaks=c(seq(-(max(allks5)), max(allks5) , by = 0.05)) titre = "allkinases SASP " mycol <- colorpanel(n=length(breaks)- titre = "allkinases SASP + p16" 1,low="green",mid="black",high="red") titre = "allkinases SASP + p16 + NFkB" #Load Normalized RT-QPCR table (do one by one) # Heatmap, "symkey = T" allks<-read.table("allkS.txt", header = TRUE, sep = "\t", dec = ",", fill = TRUE, , row.names=1 ) dev.off() allks<-read.table("allkSp16.txt", header = TRUE, sep = heatmap.2(allks5, scale = "none", col = mycol, "\t", dec = ",", fill = TRUE, , row.names=1 ) breaks=breaks, symkey = F, allks<-read.table("allkSp16NFkB.txt", header = TRUE, trace = 'none', cexRow=0.8, cexCol = 1.6, srtCol sep = "\t", dec = ",", fill = TRUE, , row.names=1 ) = 0, keysize = 1.3, -Log2 relative fold changes induction calculation key.title = "", margins = c(8, 10), # matrix of minimun values main = paste("relative FC"," ", titre), min=apply(allks, 2, min ) Rowv=F mat<-matrix(min, length(row.names(allks)),length(colnames(allks)), -corrgram byrow=TRUE) # check mat install.packages("corrgram") -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5.