Comparative Mapping Between Arabidopsis Thaliana and Brassica Nigra Indicates That Brassica Genomes Have Evolved Through Extensi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

21 CFR Ch. I (4–1–10 Edition) § 582.20
§ 582.20 21 CFR Ch. I (4–1–10 Edition) Common name Botanical name of plant source Marjoram, sweet .......................................................................... Majorana hortensis Moench. Mustard, black or brown .............................................................. Brassica nigra (L.) Koch. Mustard, brown ............................................................................ Brassica juncea (L.) Coss. Mustard, white or yellow .............................................................. Brassica hirta Moench. Nutmeg ........................................................................................ Myristica fragrans Houtt. Oregano (oreganum, Mexican oregano, Mexican sage, origan) Lippia spp. Paprika ......................................................................................... Capsicum annuum L. Parsley ......................................................................................... Petroselinum crispum (Mill.) Mansf. Pepper, black ............................................................................... Piper nigrum L. Pepper, cayenne ......................................................................... Capsicum frutescens L. or Capsicum annuum L. Pepper, red .................................................................................. Do. Pepper, white ............................................................................... Piper nigrum L. Peppermint .................................................................................. Mentha piperita L. Poppy seed -

Illinois Exotic Species List
Exotic Species in Illinois Descriptions for these exotic species in Illinois will be added to the Web page as time allows for their development. A name followed by an asterisk (*) indicates that a description for that species can currently be found on the Web site. This list does not currently name all of the exotic species in the state, but it does show many of them. It will be updated regularly with additional information. Microbes viral hemorrhagic septicemia Novirhabdovirus sp. West Nile virus Flavivirus sp. Zika virus Flavivirus sp. Fungi oak wilt Ceratocystis fagacearum chestnut blight Cryphonectria parasitica Dutch elm disease Ophiostoma novo-ulmi and Ophiostoma ulmi late blight Phytophthora infestans white-nose syndrome Pseudogymnoascus destructans butternut canker Sirococcus clavigignenti-juglandacearum Plants okra Abelmoschus esculentus velvet-leaf Abutilon theophrastii Amur maple* Acer ginnala Norway maple Acer platanoides sycamore maple Acer pseudoplatanus common yarrow* Achillea millefolium Japanese chaff flower Achyranthes japonica Russian knapweed Acroptilon repens climbing fumitory Adlumia fungosa jointed goat grass Aegilops cylindrica goutweed Aegopodium podagraria horse chestnut Aesculus hippocastanum fool’s parsley Aethusa cynapium crested wheat grass Agropyron cristatum wheat grass Agropyron desertorum corn cockle Agrostemma githago Rhode Island bent grass Agrostis capillaris tree-of-heaven* Ailanthus altissima slender hairgrass Aira caryophyllaea Geneva bugleweed Ajuga genevensis carpet bugleweed* Ajuga reptans mimosa -

Coriander Fruit. I Yield and Glucosinolate Contents of Mustard (Sinapis Sp., Brassica Sp.) Seeds
JOURNAL OF AGRICULTURAL SCIENCE IN FINLAND Maataloustieteellinen A ikakauskirja Vol. 58: 157—162, 1986 Yield and glucosinolates in mustard seeds and volatile oils in caraway seeds and coriander fruit. I Yield and glucosinolate contents of mustard (Sinapis sp., Brassica sp.) seeds 1 2 3 2 *, HÄLVÄ, S. , HIRVI, T. MÄKINEN, S. and HONKANEN, E. 1 Dept of Horticulture, University of Helsinki, SF-00710 HELSINKI, Finland 2 VTT, Food Research Laboratory, SF-02150 ESPOO, Finland 3 Dept of Nutrition, University of Helsinki, SF-00710 HELSINKI, Finland Abstract. Different varieties of yellow mustard (Sinapis alba L.), brown mustard (Bras- sica juncea (L.) Czern.) and black mustard (Brassica nigra (L.) W.D.J. Koch) were tested in 1983—1985 at three locations in Finland. The average seed yield of yellow mustard was 2220 kg/ha, it’s sinalbine content being 2.2—5.2 g/100 g. There were no major differences between the tested varieties. Varieties ‘Kirby’ and ‘Gisilba’ produced the largest yields. ‘Gisil- ba’ and ‘Ochre’ had the shortest growth periods. The sinalbine content in yellow mustard seeds varied more between the years than between the varieties. The average yield ofbrown mustard was 1620 kg/ha. The variety ‘Picra’ was slightly better than the other varieties with respect to yield and early ripening. The sinigrine content in brown mustard seeds were approximately from traces to 4.4 g/100 g those of‘Dome’, ‘Blaze’, ‘Sv 8341001’ and ‘Trowse’ being highest. Black mustard yielded less than 700 kg/ha, the sinigrine content of the seeds being 1.8—4.5 g/100g. -
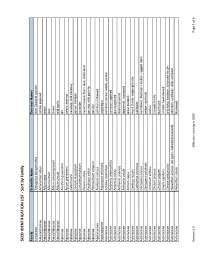
SEED IDENTIFICATION LIST - Sort by Family
SEED IDENTIFICATION LIST - Sort by Family Family Scientific Name Common Names Aizoaceae Tetragonia tetragonoides New Zealand spinach Amaranthaceae Amaranthus albus tumble pigweed Amaryllidaceae Allium cepa onion Amaryllidaceae Allium porrum leek Amaryllidaceae Allium schoenoprasum chives Amaryllidaceae Allium vineale wild garlic Apiaceae Anethum graveolens dill Apiaceae Apium graveolens celery, celeriac Apiaceae Carum carvi caraway; wild caraway Apiaceae Conium maculatum poison hemlock Apiaceae Coriandrum sativum coriander Apiaceae Daucus carota carrot; Queen Ane's lace; wild carrot Apiaceae Pastinaca sativa parsnip; wild parsnip Apiaceae Petroselinum crispum parsley Apocynaceae Asclepias syriaca common milkweed Asparagaceae Asparagus officinalis asparagus Asteraceae Achillea millefolium common yarrow, woolly yarrow Asteraceae Ambrosia artemisiifolia common ragweed Asteraceae Ambrosia trifida giant ragweed Asteraceae Anthemis arvensis field chamomile Asteraceae Anthemis cotula dogfennel, mayweed Asteraceae Arctium lappa great burdock Asteraceae Carduus nutans musk thistle, nodding thistle Asteraceae Carthamus tinctorius safflower Asteraceae Centaurea cyanus cornflower, bachelor's button, ragged robin Asteraceae Centaurea solstitialis yellow starthistle Asteraceae Cichorium endivia endive Asteraceae Cirsium arvense Canada thistle Asteraceae Cirsium vulgare bull thistle Asteraceae Crepis capillaris smooth hawksbeard Asteraceae Cynara cardunculus artichoke, cardoon, artichoke thistle Asteraceae Helianthus annuus (all types, cultivated and -

Antibacterial Activity of Different Plant and Callus Extracts a Comparative Study
INTERNATIONAL JOURNAL OF SCIENTIFIC & TECHNOLOGY RESEARCH VOLUME 2, ISSUE 10, OCTOBER 2013 ISSN 2277-8616 Antibacterial Activity Of Different Plant And Callus Extracts A Comparative Study V.D. Jadhav, S. M. Bhanuwanshe, S. P. Patil, D.V. Chaudhari, M.B. Adke Abstract: Current study includes antibacterial activity of different plant and callus extract such as Momordica charantia (Karela), Cucurbita pepo (Pumpkin), Capsicum annum (Chilli), Coriander sativum (Coriander), Brassica nigra (Mustard), Nigella sativa (Black Cumin), Trigonella foenum (Fenugreek) against eight pathogenic bacteria of which five are gram positive such as Staphylococcus aureus, Bacillus cereus, Bacillus subtilis, Streptococcus sp, Bacillus megaterium and three are gram negative such as Escherichia coli, Proteus vulgaris, Pseudomonas aeruginosa. Plant materials were extracted using ethanol. All the extracts showed zone of inhibition against these pathogenic bacteria which were tested by agar well diffusion method. The leaf extract of Momordica charantia, Nigella sativa, Barassica nigra showed good zone of inhibition as compared to other extracts against S.aureus (13mm, 17mm,11mm), E.coli (13mm,10mm,18mm), P. aeruginosa (11mm,12mm,10mm), Streptocococcus (14mm, 15mm, 10mm), B.subtilis (10mm,17mm,10mm), P. vulgaris (15mm,18mm, 10mm), B.cereus (13mm,16mm,12mm), B.megaterium (12mm,15mm,10mm). Explants from in vitro grown plants were cultured on MS-Medium with different combination of IAA and Kinetin for callus induction. Among those combination of IAA and Kinetin (0.1 x 0.0), (1.0 x 0.0), (0.4 x 0.5), (1.0 x 0.5), (1.5 x 0.5), (0.4 x 1.0), (0.8 x1.0), (0.1 x 1.5) mg/L have showed maximum growth of calli. -

Brassica Nigra and Curcuma Longa Compounds Affecting Interactions Between Spodoptera Exigua and Its Natural Enemies Cotesia Flav
Original Article Dose-Response: An International Journal January-March 2019:1-10 Brassica nigra and Curcuma longa ª The Author(s) 2019 Article reuse guidelines: Compounds Affecting Interactions Between sagepub.com/journals-permissions DOI: 10.1177/1559325819827454 Spodoptera exigua and Its Natural Enemies journals.sagepub.com/home/dos Cotesia flavipes and Podisus maculiventris Wagner de Souza Tavares1,2,3, Jesusa Crisostomo Legaspi2, Ancideriton´ Antonio de Castro2,3,4, Hany Ahmed Fouad4,5, Muhammad Haseeb3, Robert L. Meagher, Jr6, Lambert H.B. Kanga3, and Jose´ Cola Zanuncio4 Abstract The interaction Spodoptera exigua Hu¨bner (Lepidoptera: Noctuidae) Â its natural enemies Cotesia flavipes Cameron (Hyme- noptera: Braconidae) and Podisus maculiventris Say (Heteroptera: Pentatomidae) Â botanical compounds with and without synergist is unknown; therefore, it was studied under controlled conditions. The objective of this study was to evaluate the direct mortality of P. maculiventris nymphs and adults and indirect by this predator feeding on S. exigua larvae treated after being exposed to parasitism by C. flavipes. Brassica nigra L. (Brassicales: Brassicaceae) and Curcuma longa L. (Zingiberales: Zingiberaceae) com- pounds, with and without lead (II) oxide (PbO), were tested as insecticides. The mortality of first and second instars P. maculi- ventris was high with turmeric essential oil by topical application. The PbO increased the predator mortality in combination with turmeric powder, crude essential oil, and ar-turmerone. This last derivative caused also the highest mortality of P. maculiventris nymphs when ingested through treated S. exigua larvae that were previously subjected to parasitism. Turmeric powder and its derivatives, with and without PbO, should not be used in areas with P. -

Chemical Composition and Antimicrobial Activity in Cold Press Oil of Fennel, Anise, White and Black Mustard Seeds
Original Article Chemical Composition and Antimicrobial Activity in Cold Press Oil of Fennel, Anise, White and Black Mustard Seeds Çağrı Olgun1, Osman Emre Özkan1, Betül Güney2, Esma Sena Pattabanoglu3, Kerim Güney3, Mahmut Gür1 1Department of Forest Industrial Engineering, Faculty of Forestry, Kastamonu University, TURKEY 2Graduate School of Natural and Applied Sciences, Department of Biology, Ankara University, TURKEY 3Department of Forest Engineering, Faculty of Forestry, Kastamonu University, TURKEY ABSTRACT In this study, the cold press oil components and antimicrobial activities of fennel (Foeniculum vulgare) and anise (Pimpinella anisum) and white mustard (Sinapis alba) and black mustard (Brassica nigra) species seeds, which are widely used by the people for alternative medicine, were determined. F. vulgare, P. anisum, S. alba and B. nigra species seeds were obtained from cultivated areas in central Anatolia in Turkey. The oil was extracted by using a screw press (MP-001 Cold Press, Turkey), and the volatile oil components and fatty acid components in these oils were analysed by GCMS and total phenolic content, total flavonoid content and antioxidant activities by DPPH and FRAP (%) method were determined. Antimicrobial activities of obtained oils were investigated by using minimum inhibitory concentration (MIC) test by against 18 different species microorganisms. In the GCMS results, F. vulgare and P. anisum oils were found to be the most abundant components which were anethole (89.74%, 88.95%, respectively). According to these results, the plants oils didn’t show any antimicrobial activities against tested microorganisms. However especially white and black mustard oils showed strong antioxidant activity when compared with artificial antioxidants. Keywords: Cold Press Oils, Gs-Mc, Antioxidant Activity and Antimicrobial Activities. -

Table E-1. Vegetation Species Found on Wake Atoll
Table E-1. Vegetation Species Found on Wake Atoll Scientific Name Common Name Abutilon albescens Sweet monkeybush Abutilon asiaticum var. albescens Indian mallow Agave americana American century plant Agave angustifolia century plant Agave sisalana Sisal Agave sp. agave sp. Aglaonema commutatum Aglaonema Allium cepa Onion Allium fistulosum Green onion Allium sp. Onion sp. Allium tuberosum Chinese chive Aloe vera Aloe Alpinia galanga Greater galangal Alpinia purpurata Pink ginger; Jungle Queen Amaranthus dubius Spleen amaranth Amaranthus graecizans Tumbleweed Amaranthus tricolor Joseph′s coat Amaranthus viridis Slender amaranth Ananas comosus Pineapple Anethum graveolens Dill Annona muricata Soursop Annona squamosa Sweetsop Apium petroselinum Garden parsley Araucaria heterophylla Norfolk Island pine Asparagus densiflorus Sprenger asparagus fern Asplenium nidus Bird’s-nest fern Barringtonia asiatica Fish poison tree Bauhinia sp. Camel’s foot tree Bidens alba white beggar-ticks Bidens pilosa var. minor Beggar-ticks Boerhavia albiflora var. powelliae -- Boerhavia diffusa Red Spiderling Boerhavia repens anena Boerhavia sp. Spiderling sp. Bothriochloa pertusa Indian blue grass Bougainvillea spectabilis bougainvillea Brassica nigra Mustard Brassica oleracea var. italica Brocolli Caesalpinia bonduc Grey nickers Caladium bicolor Caladium Calotropis gigantea Crown flower Capsicum frutescens Cayenne pepper Capsicum annuum chili pepper Table E-1. Vegetation Species Found on Wake Atoll Scientific Name Common Name Carica papaya Papaya Casuarina equisetifolia -

UNIVERSITY of CALIFORNIA, SAN DIEGO the Dynamics Driving the Invasion of Brassica Nigra and Foeniculum Vulgare in California
UNIVERSITY OF CALIFORNIA, SAN DIEGO The dynamics driving the invasion of Brassica nigra and Foeniculum vulgare in California: the interaction of competition, herbivory, and seed source A Thesis submitted in partial satisfaction of the requirements for the degree Master of Science in Biology by Victoria Anderson Hanna Committee in charge: Professor Carolyn M. Kurle, Chair Professor Elsa Cleland Professor Joshua Kohn 2014 The Thesis of Victoria Anderson Hanna is approved and it is acceptable in quality and form for publication on microfilm and electronically: _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ Chair University of California, San Diego 2014 iii TABLE OF CONTENTS Signature Page.................................................................................... iii Table of Contents............................................................................... iv List of Tables...................................................................................... v List of Figures..................................................................................... vii Abstract of the Thesis......................................................................... iv Introduction........................................................................................ 1 Methods.............................................................................................. 5 -

BIOL 317: Plant Identification and Classification Summer 2015 - Notes
BIOL 317: Plant Identification and Classification Summer 2015 - Notes Week 6 – Tuesday Plant reproduction and breeding systems(cont.) Advantages of different breeding systems • asexual reproduction and selfing . reproductive assurance - can reproduce even in small, isolated populations . reduced reproductive effort - no need to produce showy flowers or flowers at all . locally adapted offspring in stable environments . 100% of genes passed to offspring • outcrossing . greater genetic variability in offspring - especially useful in changing environments . reduces inbreeding depression - maintenance of heterozygosity hides expression of deleterious recessive alleles Myrtales other important families include Lythraceae (loosestrife, crape myrtle, pomegranate) and Myrtaceae (eucalyptus, guava, allspice) Onagraceae (evening-primrose family) 22 genera, 656 spp. distributed widely, but especially diverse in western North America and South America herbaceous or woody (shrubs) leaves - simple; variously arranged flowers • actinomorphic or zygomorphic • hypanthium common • sepals usually 4 • petals usually 4 • stamens 4 or 8 . pollen held in strands by sticky viscin threads • pistil compound (4 carpels); ovary inferior • fruit: capsule or berry includes PNW wildflowers (Chamerion angustifolium - fireweed, Oenothera spp. - evening-primrose, Epilobium spp. - willowherb, Clarkia spp., etc.) and ornamentals (Fuchsia spp. - fuchsia, Oenothera spp., Epilobium spp., etc.) Brassicales other important families include Capparidaceae (caper; -

(Brassica Nigra) Extract on the Prefrontal Cortex of Adult Wistar Rats
Original Article www.anatomy.org.tr Received: August 13, 2016; Accepted: September 2, 2017 doi:10.2399/ana.17.027 Dose-dependent effect of black mustard seeds (Brassica nigra) extract on the prefrontal cortex of adult Wistar rats Tolulope Timothy Arogundade1,2, Bernard Ufuoma Enaibe1,2, Oluwaseun Olaniyi Adigun2, Foyeke Munirat Adigun2, Ismail Temitayo Gbadamosi2 1Department of Anatomy, Faculty of Basic Medical Sciences, Adeleke University, Ede, Nigeria 2Department of Anatomy, Faculty of Basic Medical Sciences, University of Ilorin, Ilorin, Nigeria Abstract Objectives: Mustard seeds, apart from being a culinary essential, have had medicinal applications dating back to the time of Hippocrates. It has in fact been once mentioned as the greatest herb ever. We explored the dose dependent effects of the crude aqueous extract of Brassica nigra (Black mustard seeds) on the prefrontal cortex of adult Wistar rats. Methods: 20 adult female rats weighing an average of 180±20 g were used. They were split into 4 groups (n=5); Group A (received extract at 200 mg/kg body weight), Group B (received extract at 100 mg/kg body weight), Group C (received extract at 50 mg/kg body weight), and Group D (received distilled water ad libitum). All of the animals were subjected to the Y-maze spontaneous alternation test for neurobehavioural analyses following 28-day administration of the extracts. Animals were sacri- ficed 24 hours after taking the last day of administration. Results: Our results showed that neurobehavioural analyses are significantly hampered in animals receiving 200 mg/kg extract in comparison to the control group. In treatment groups, increased dose of extract elevated the level of MDA, but reduced the level of SOD. -

Brassica Spp.) – 151
II.3. BRASSICA CROPS (BRASSICA SPP.) – 151 Chapter 3. Brassica crops (Brassica spp.) This chapter deals with the biology of Brassica species which comprise oilseed rape, turnip rape, mustards, cabbages and other oilseed crops. The chapter contains information for use during the risk/safety regulatory assessment of genetically engineered varieties intended to be grown in the environment (biosafety). It includes elements of taxonomy for a range of Brassica species, their centres of origin and distribution, reproductive biology, genetics, hybridisation and introgression, crop production, interactions with other organisms, pests and pathogens, breeding methods and biotechnological developments, and an annex on common pathogens and pests. The OECD gratefully acknowledges the contribution of Dr. R.K. Downey (Canada), the primary author, without whom this chapter could not have been written. The chapter was prepared by the OECD Working Group on the Harmonisation of Regulatory Oversight in Biotechnology, with Canada as the lead country. It updates and completes the original publication on the biology of Brassica napus issued in 1997, and was initially issued in December 2012. Data from USDA Foreign Agricultural Service and FAOSTAT have been updated. SAFETY ASSESSMENT OF TRANSGENIC ORGANISMS: OECD CONSENSUS DOCUMENTS, VOLUME 5 © OECD 2016 152 – II.3. BRASSICA CROPS (BRASSICA SPP.) Introduction The plants within the family Brassicaceae constitute one of the world’s most economically important plant groups. They range from noxious weeds to leaf and root vegetables to oilseed and condiment crops. The cole vegetables are perhaps the best known group. Indeed, the Brassica vegetables are a dietary staple in every part of the world with the possible exception of the tropics.