Molecular Cloning and Expression Profile of Class E Genes Related To
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Survey of Roadside Alien Plants in Hawai`I Volcanoes National Park and Adjacent Residential Areas 2001–2005
Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANts IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 Linda W. Pratt1 Keali`i F. Bio2 James D. Jacobi1 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kilauea Field Station, P.O. Box 44, Hawaii National Park, HI 96718 2 Hawai‘i Cooperative Studies Unit, University of Hawai‘i at Hilo, P.O. Box 44, Hawai‘i National Park, HI 96718 Hawai‘i Cooperative Studies Unit University of Hawai‘i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This product was prepared under Cooperative Agreement CA03WRAG0036 for the Pacific Island Ecosystems Research Center of the U.S. Geological Survey. Technical Report HCSU-032 SURVEY OF ROADSIDE ALIEN PLANTS IN HAWAI`I VOLCANOES NATIONAL PARK AND ADJACENT RESIDENTIAL AREAS 2001–2005 1 2 1 LINDA W. PRATT , KEALI`I F. BIO , AND JAMES D. JACOBI 1 U.S. Geological Survey, Pacific Island Ecosystems Research Center, Kīlauea Field Station, P.O. Box 44, Hawai`i Volcanoes National Park, HI 96718 2 Hawaii Cooperative Studies Unit, University of Hawai`i at Hilo, Hilo, HI 96720 Hawai`i Cooperative Studies Unit University of Hawai`i at Hilo 200 W. Kawili St. Hilo, HI 96720 (808) 933-0706 September 2012 This article has been peer reviewed and approved for publication consistent with USGS Fundamental Science Practices ( http://pubs.usgs.gov/circ/1367/ ). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. -

The Developmental and Genetic Bases of Apetaly in Bocconia Frutescens
Arango‑Ocampo et al. EvoDevo (2016) 7:16 DOI 10.1186/s13227-016-0054-6 EvoDevo RESEARCH Open Access The developmental and genetic bases of apetaly in Bocconia frutescens (Chelidonieae: Papaveraceae) Cristina Arango‑Ocampo1, Favio González2, Juan Fernando Alzate3 and Natalia Pabón‑Mora1* Abstract Background: Bocconia and Macleaya are the only genera of the poppy family (Papaveraceae) lacking petals; how‑ ever, the developmental and genetic processes underlying such evolutionary shift have not yet been studied. Results: We studied floral development in two species of petal-less poppies Bocconia frutescens and Macleaya cordata as well as in the closely related petal-bearing Stylophorum diphyllum. We generated a floral transcriptome of B. frutescens to identify MADS-box ABCE floral organ identity genes expressed during early floral development. We performed phylogenetic analyses of these genes across Ranunculales as well as RT-PCR and qRT-PCR to assess loci- specific expression patterns. We found that petal-to-stamen homeosis in petal-less poppies occurs through distinct developmental pathways. Transcriptomic analyses of B. frutescens floral buds showed that homologs of all MADS-box genes are expressed except for the APETALA3-3 ortholog. Species-specific duplications of other ABCE genes inB. frute- scens have resulted in functional copies with expanded expression patterns than those predicted by the model. Conclusions: Petal loss in B. frutescens is likely associated with the lack of expression of AP3-3 and an expanded expression of AGAMOUS. The genetic basis of petal identity is conserved in Ranunculaceae and Papaveraceae although they have different number of AP3 paralogs and exhibit dissimilar floral groundplans. -

David A. Rasmussen, 2 Elena M. Kramer, 3 and Elizabeth A. Zimmer 4
American Journal of Botany 96(1): 96–109. 2009. O NE SIZE FITS ALL? M OLECULAR EVIDENCE FOR A COMMONLY INHERITED PETAL IDENTITY PROGRAM IN RANUNCULALES 1 David A. Rasmussen, 2 Elena M. Kramer, 3 and Elizabeth A. Zimmer 4 Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, Massachusetts 02138 USA Petaloid organs are a major component of the fl oral diversity observed across nearly all major clades of angiosperms. The vari- able morphology and development of these organs has led to the hypothesis that they are not homologous but, rather, have evolved multiple times. A particularly notable example of petal diversity, and potential homoplasy, is found within the order Ranunculales, exemplifi ed by families such as Ranunculaceae, Berberidaceae, and Papaveraceae. To investigate the molecular basis of petal identity in Ranunculales, we used a combination of molecular phylogenetics and gene expression analysis to characterize APETALA3 (AP3 ) and PISTILLATA (PI ) homologs from a total of 13 representative genera of the order. One of the most striking results of this study is that expression of orthologs of a single AP3 lineage is consistently petal-specifi c across both Ranunculaceae and Berberidaceae. We conclude from this fi nding that these supposedly homoplastic petals in fact share a developmental genetic program that appears to have been present in the common ancestor of the two families. We discuss the implications of this type of molecular data for long-held typological defi nitions of petals and, more broadly, the evolution of petaloid organs across the angiosperms. Key words: APETALA3 ; MADS box genes; petal evolution; PISTILLATA ; Ranunculales. -

Reconstructing the Basal Angiosperm Phylogeny: Evaluating Information Content of Mitochondrial Genes
55 (4) • November 2006: 837–856 Qiu & al. • Basal angiosperm phylogeny Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes Yin-Long Qiu1, Libo Li, Tory A. Hendry, Ruiqi Li, David W. Taylor, Michael J. Issa, Alexander J. Ronen, Mona L. Vekaria & Adam M. White 1Department of Ecology & Evolutionary Biology, The University Herbarium, University of Michigan, Ann Arbor, Michigan 48109-1048, U.S.A. [email protected] (author for correspondence). Three mitochondrial (atp1, matR, nad5), four chloroplast (atpB, matK, rbcL, rpoC2), and one nuclear (18S) genes from 162 seed plants, representing all major lineages of gymnosperms and angiosperms, were analyzed together in a supermatrix or in various partitions using likelihood and parsimony methods. The results show that Amborella + Nymphaeales together constitute the first diverging lineage of angiosperms, and that the topology of Amborella alone being sister to all other angiosperms likely represents a local long branch attrac- tion artifact. The monophyly of magnoliids, as well as sister relationships between Magnoliales and Laurales, and between Canellales and Piperales, are all strongly supported. The sister relationship to eudicots of Ceratophyllum is not strongly supported by this study; instead a placement of the genus with Chloranthaceae receives moderate support in the mitochondrial gene analyses. Relationships among magnoliids, monocots, and eudicots remain unresolved. Direct comparisons of analytic results from several data partitions with or without RNA editing sites show that in multigene analyses, RNA editing has no effect on well supported rela- tionships, but minor effect on weakly supported ones. Finally, comparisons of results from separate analyses of mitochondrial and chloroplast genes demonstrate that mitochondrial genes, with overall slower rates of sub- stitution than chloroplast genes, are informative phylogenetic markers, and are particularly suitable for resolv- ing deep relationships. -

HAWAII and SOUTH PACIFIC ISLANDS REGION - 2016 NWPL FINAL RATINGS U.S
HAWAII and SOUTH PACIFIC ISLANDS REGION - 2016 NWPL FINAL RATINGS U.S. ARMY CORPS OF ENGINEERS, COLD REGIONS RESEARCH AND ENGINEERING LABORATORY (CRREL) - 2013 Ratings Lichvar, R.W. 2016. The National Wetland Plant List: 2016 wetland ratings. User Notes: 1) Plant species not listed are considered UPL for wetland delineation purposes. 2) A few UPL species are listed because they are rated FACU or wetter in at least one Corps region. Scientific Name Common Name Hawaii Status South Pacific Agrostis canina FACU Velvet Bent Islands Status Agrostis capillaris UPL Colonial Bent Abelmoschus moschatus FAC Musk Okra Agrostis exarata FACW Spiked Bent Abildgaardia ovata FACW Flat-Spike Sedge Agrostis hyemalis FAC Winter Bent Abrus precatorius FAC UPL Rosary-Pea Agrostis sandwicensis FACU Hawaii Bent Abutilon auritum FACU Asian Agrostis stolonifera FACU Spreading Bent Indian-Mallow Ailanthus altissima FACU Tree-of-Heaven Abutilon indicum FAC FACU Monkeybush Aira caryophyllea FACU Common Acacia confusa FACU Small Philippine Silver-Hair Grass Wattle Albizia lebbeck FACU Woman's-Tongue Acaena exigua OBL Liliwai Aleurites moluccanus FACU Indian-Walnut Acalypha amentacea FACU Alocasia cucullata FACU Chinese Taro Match-Me-If-You-Can Alocasia macrorrhizos FAC Giant Taro Acalypha poiretii UPL Poiret's Alpinia purpurata FACU Red-Ginger Copperleaf Alpinia zerumbet FACU Shellplant Acanthocereus tetragonus UPL Triangle Cactus Alternanthera ficoidea FACU Sanguinaria Achillea millefolium UPL Common Yarrow Alternanthera sessilis FAC FACW Sessile Joyweed Achyranthes -

Title Insect-Flower Relationship in the Temperate Deciduous Forest Of
Insect-flower Relationship in the Temperate Deciduous Forest Title of Kibune, Kyoto : An Overview of the Flowering Phenology and the Seasonal Pattern of Insect Visits INOUE, Tamiji; KATO, Makoto; KAKUTANI, Takehiko; Author(s) SUKA, Takeshi; ITINO, Takao Contributions from the Biological Laboratory, Kyoto Citation University (1990), 27(4): 377-464 Issue Date 1990-08-20 URL http://hdl.handle.net/2433/156100 Right Type Departmental Bulletin Paper Textversion publisher Kyoto University Contr. biol, Lab. Kyoto Univ,, Vol. 27, pp. 377-463 Issued 20 August 1990 Insect-flower Relationship in the Temperate Deciduous 'Forest of Kibune, Kyoto: An Overview of the Flowering Phenology and the Seasonal Pattern of Insect Visits' Tamiji INouE, Makoto KATo, Takehiko KAKuTANi, Takeshi SuKA and Takao IT[No ABSTRACT In 1984 -1987, insect visitors to fiowers werebimonthly or weekly surveyed on a total of 115 plant species or 49 families in the temperate deciduous forest of Kibune, Kyoto, Japan. Flowering was observed from early April to early November, The number of plant species that concurrently bloomed was nine to 17 from May to September. Themonthly total number of flowering plant species peaked twice in May (34 spp.) and September (33 spp,). From April to August, floweringperiods werestaggered arnong congeneric woody species, e.g., Lindera, Rubus, Hydrangea and Deutzia. A total of 4603 individuals of 889 species in 12 orders of Insecta and 2 orders of Arachnoidea were collected. The most abundant order was Hymeno- ptera (46 O/o of the total number of individuals), and it was followed by Diptera (30 O/o) and Coleoptera (140/o). -

9:00 Am PLACE
CARTY S. CHANG INTERIM CHAIRPERSON DAVID Y. IGE BOARD OF LAND AND NATURAL RESOURCES GOVERNOR OF HAWAII COMMISSION ON WATER RESOURCE MANAGEMENT KEKOA KALUHIWA FIRST DEPUTY W. ROY HARDY ACTING DEPUTY DIRECTOR – WATER AQUATIC RESOURCES BOATING AND OCEAN RECREATION BUREAU OF CONVEYANCES COMMISSION ON WATER RESOURCE MANAGEMENT STATE OF HAWAII CONSERVATION AND COASTAL LANDS CONSERVATION AND RESOURCES ENFORCEMENT DEPARTMENT OF LAND AND NATURAL RESOURCES ENGINEERING FORESTRY AND WILDLIFE HISTORIC PRESERVATION POST OFFICE BOX 621 KAHOOLAWE ISLAND RESERVE COMMISSION LAND HONOLULU, HAWAII 96809 STATE PARKS NATURAL AREA RESERVES SYSTEM COMMISSION MEETING DATE: April 27, 2015 TIME: 9:00 a.m. PLACE: Department of Land and Natural Resources Boardroom, Kalanimoku Building, 1151 Punchbowl Street, Room 132, Honolulu. AGENDA ITEM 1. Call to order, introductions, move-ups. ITEM 2. Approval of the Minutes of the June 9, 2014 N atural Area Reserves System Commission Meeting. ITEM 3. Natural Area Partnership Program (NAPP). ITEM 3.a. Recommendation to the Board of Land and Natural Resources approval for authorization of funding for The Nature Conservancy of Hawaii for $663,600 during FY 16-21 for continued enrollment in the natural area partnership program and acceptance and approval of the Kapunakea Preserve Long Range Management Plan, TMK 4-4-7:01, 4-4-7:03, Lahaina, Maui. ITEM 3.b. Recommendation to the Board of Land and Natural Resources approval for authorization of funding for The Nature Conservancy of Hawaii for $470,802 during FY 16-21 for continued enrollment in the natural area partnership program and acceptance and approval of the Pelekunu Long Range Management Plan, TMK 5-4- 3:32, 5-9-6:11, Molokai. -

Towards a Phylogenetic Nomenclature of Tracheophyta
Cantino & al. • Phylogenetic nomenclature of Tracheophyta TAXON 56 (3) • August 2007: 822–846 PHYLOGENEtic noMEncLAturE Towards a phylogenetic nomenclature of Tracheophyta Philip D. Cantino2, James A. Doyle1,3, Sean W. Graham1,4, Walter S. Judd1,5, Richard G. Olmstead1,6, Douglas E. Soltis1,5, Pamela S. Soltis1,7 & Michael J. Donoghue8 1 Authors are listed alphabetically, except for the first and last authors. 2 Department of Environmental and Plant Biology, Ohio University, Athens, Ohio 45701, U.S.A. [email protected] (author for correspondence) 3 Section of Evolution and Ecology, University of California, Davis, California 95616, U.S.A. 4 UBC Botanical Garden and Centre for Plant Research, 6804 SW Marine Drive, The University of British Columbia, Vancouver, British Columbia, Canada V6T 1Z4 5 Department of Botany, University of Florida, Gainesville, Florida 32611-8526, U.S.A. 6 Department of Biology, P.O. Box 355325, University of Washington, Seattle, Washington 98195-5325, U.S.A. 7 Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611, U.S.A. 8 Department of Ecology & Evolutionary Biology and Peabody Museum of Natural History, Yale University, P.O. Box 208106, New Haven, Connecticut 06520-8106, U.S.A. This is an abbreviated version of a paper that appears in full in the Electronic supplement to Taxon. Phylogenetic definitions are provided for the names of 20 clades of vascular plants (plus 33 others in the electronic supple- ment). Emphasis has been placed on well-supported clades that are widely known to non-specialists and/or have a deep origin within Tracheophyta or Angiospermae. -

Field Instructions for the Periodic Inventory of The
FIELD INSTRUCTIONS FOR THE PERIODIC INVENTORY OF THE COMMONWEALTH OF THE NORTHERN MARIANA ISLANDS 2015 FOREST INVENTORY AND ANALYSIS RESOURCE MONITORING AND ASSESSMENT PROGRAM PACIFIC NORTHWEST RESEARCH STATION USDA FOREST SERVICE THIS MANUAL IS BASED ON: FOREST INVENTORY AND ANALYSIS NATIONAL CORE FIELD GUIDE VOLUME I: FIELD DATA COLLECTION PROCEDURES VERSION 6.1 Cover image by Gretchen Bracher pg.I Table of Contents CHAPTER 1 INTRODUCTION . 15 SECTION 1.1 ORGANIZATION OF THIS MANUAL. 15 SECTION 1.2 THE INVENTORY. 16 SECTION 1.3 PRODUCTS . 16 SECTION 1.4 UNITS OF MEASURE . 16 SECTION 1.5 PLOT DESIGN GENERAL DESCRIPTION . 16 SUBSECTION 1.5.1 PLOT LAYOUT . .17 SUBSECTION 1.5.2 DATA ARE COLLECTED ON PLOTS AT THE FOLLOWING LEVELS .17 SECTION 1.6 QUALITY ASSURANCE/QUALITY CONTROL . 18 SUBSECTION 1.6.1 GENERAL DESCRIPTION . .18 SECTION 1.7 SAFETY . 18 SUBSECTION 1.7.1 SAFETY IN THE WOODS. .18 SUBSECTION 1.7.2 SAFETY ON THE ROAD . .19 SUBSECTION 1.7.3 WHAT TO DO IF INJURED. .19 CHAPTER 2 LOCATING THE PLOT . 21 SECTION 2.1 LOCATING AN ESTABLISHED PLOT . 21 SUBSECTION 2.1.1 NAVIGATING WITH PHOTOGRAPHY. .21 SUBSECTION 2.1.2 NAVIGATING WITH GPS . .21 SUBSECTION 2.1.3 NAVIGATING WITH REFERENCE POINT (RP) DATA . .22 SUBSECTION 2.1.4 REVERSE REFERENCE POINT (RP) METHOD . .22 SECTION 2.2 ESTABLISHED PLOT ISSUES . 22 SUBSECTION 2.2.1 DIFFICULTY FINDING ESTABLISHED PLOTS. 22 SUBSECTION 2.2.2 INCORRECTLY INSTALLED PLOT . .23 SUBSECTION 2.2.3 INCORRECTLY INSTALLED SUBPLOT OR MICROPLOT. .23 SUBSECTION 2.2.4 PC STAKE OR SUBPLOT/MICROPLOT PIN MISSING OR MOVED . -
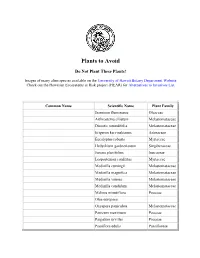
Plants to Avoid
Plants to Avoid Do ot Plant These Plants! Images of many alien species available on the University of Hawaii Botany Department Website Check out the Hawaiian Ecosystems at Risk project (HEAR) for Alternatives to Invasives List Common ame Scientific ame Plant Family Jasminim fluminense Oleaceae Arthrostema ciliatum Melastomataceae Dissotis rotundifolia Melastomataceae Erigeron karvinskianus Asteraceae Eucalyptus robusta Myrtaceae Hedychium gardnerianum Singiberaceae Juncus planifolius Juncaceae Lospostemon confertus Myrtaceae Medinilla cumingii Melastomataceae Medinilla magnifica Melastomataceae Medinilla venosa Melastomataceae Medinilla candidum Melastomataceae Melinis minutiflora Poaceae Olea europaea Oxyspora paniculata Melastomataceae Panicum maximum Poaceae Paspalum urvillei Poaceae Passiflora edulis Passifloreae Phormium tenax Agavaceae Pinus taeda Pinaceae Prosopis pallida Favaceae Pterolepis glomerata Melastomataceae Rhodomyrtus tomentosa Myrtaceae Schefflera actinophylla Araliaceae Syzygium jambos Myrtaceae Australian blackwood Acacia melanoxylon Mimosaceae Australian tree fern Cyathea cooperi Cyatheaceae Australian tree fern Sphaeropteris cooperi Cyatheaceae Beggar's tick, Spanish needle Bidens pilosa Asteraceae California grass Brachiaria mutica Poaceae Chinese banyan, Malayan banyan Ficus mirocarpa Moraceae Chinese violet Asystasia gangetica Acanthaceae Christmasberry, Brazilian pepper Schinus terebinthifolius Anacardiaceae Formosan koa Acacia confusa Mimosaceae German ivy Senecio mikanioides Asteraceae Japanese honeysuckle -

Street Tree Inventory Report Grant Park Neighborhood October 2016 Street Tree Inventory Report: Grant Park Neighborhood October 2016
Street Tree Inventory Report Grant Park Neighborhood October 2016 Street Tree Inventory Report: Grant Park Neighborhood October 2016 Written by: Kat Davidson, Angie DiSalvo, Julie Fukuda, Jim Gersbach, Jeremy Grotbo, and Jeff Ramsey Portland Parks & Recreation Urban Forestry 503-823-4484 [email protected] http://portlandoregon.gov/parks/treeinventory Grant Park Tree Inventory Organizers: Liz Hay Staff Neighborhood Coordinator: Jim Gersbach Data Collection Volunteers: Linda Brannan, Doug Brazil, Neff Breen, Rick Burkard, Patrick Burns, Dianna Choi, Catherine Clark, Don Crossley, Ann DeNies, Mary Desch, Russell Eng, Gregg Everhart, Claudia Fabbrini, Karla Fitzwater, Liz Hay, Leo Helm, Pamela Hickman, Lisa Horowitz, Martha Irvine, Kiel Jenkins, Ben Jones, James Keiter, Bill Kownacki, Fred Kratz, Marc Langhammer, Kate Laudermilk, Ariel Lewin, Louis Miles, Melinda Moeur, Shelley Morrison, Tamara Olcott, Larry Rabinowitz, Bruce Richard, Kyna Rubin, Amy Simpson, Matt Vellella, Aaron Wolf, and Samantha Wolf Data Entry Volunteers: Max Blasdel, Michael Brehm, Dylan Eglin, Tiffany Eurich, Spencer Keller, Matthew Pryzborski, Nathan Riggsby, Takayuki Shigematsu, Joshua Sindel, Rebecca Tait, and Shauna Volk Arborist-on-Call Volunteers: Van Bogner, Casey Clapp, and Fred Nilsen GIS Technical Support: Josh Darling, Portland Parks & Recreation Financial Support: Portland Parks & Recreation Cover Photos (from top left to bottom right): 1) Unusual burgundy seeds on a Euptelea pleiosperma, an extremely rare find in Portland. 2) The changing leaves of a Persian ironwood (Parrotia persica). 3) A pair of katsuras (Cercidiphyllum japonicum) beginning to show fall color. 4) Chains of seeds forming on a sourwood (Oxydendrum arboreum). 5) The fruit of a dove tree (Davidia involucrata). 6) Flaking bark on a mature redbud (Cercis canadensis). -

LETTER Doi:10.1038/Nature12872
LETTER doi:10.1038/nature12872 Three keys to the radiation of angiosperms into freezing environments Amy E. Zanne1,2, David C. Tank3,4, William K. Cornwell5,6, Jonathan M. Eastman3,4, Stephen A. Smith7, Richard G. FitzJohn8,9, Daniel J. McGlinn10, Brian C. O’Meara11, Angela T. Moles6, Peter B. Reich12,13, Dana L. Royer14, Douglas E. Soltis15,16,17, Peter F. Stevens18, Mark Westoby9, Ian J. Wright9, Lonnie Aarssen19, Robert I. Bertin20, Andre Calaminus15, Rafae¨l Govaerts21, Frank Hemmings6, Michelle R. Leishman9, Jacek Oleksyn12,22, Pamela S. Soltis16,17, Nathan G. Swenson23, Laura Warman6,24 & Jeremy M. Beaulieu25 Early flowering plants are thought to have been woody species to greater heights: as path lengths increase so too does resistance5. restricted to warm habitats1–3. This lineage has since radiated into Among extant strategies, the most efficient method of water delivery almost every climate, with manifold growth forms4. As angiosperms is through large-diameter water-conducting conduits (that is, vessels spread and climate changed, they evolved mechanisms to cope with and tracheids) within xylem5. episodic freezing. To explore the evolution of traits underpinning Early in angiosperm evolution they probably evolved larger conduits the ability to persist in freezing conditions, we assembled a large for water transport, especially compared with their gymnosperm cousins14. species-level database of growth habit (woody or herbaceous; 49,064 Although efficient in delivering water, these larger cells would have species), as well as leaf phenology (evergreen or deciduous), diameter impeded angiosperm colonization of regions characterized by episodic of hydraulic conduits (that is, xylem vessels and tracheids) and climate freezing14,15, as the propensity for freezing-induced embolisms (air bub- occupancies (exposure to freezing).