Classifications and Other Ordering Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Selection, Constraint, and Adaptation in the Visual Genes of Neotropical Cichlid Fishes and Other Vertebrates
SELECTION, CONSTRAINT, AND ADAPTATION IN THE VISUAL GENES OF NEOTROPICAL CICHLID FISHES AND OTHER VERTEBRATES by Frances Elisabeth Hauser A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Ecology and Evolutionary Biology University of Toronto © Copyright by Frances E. Hauser 2018 SELECTION, CONSTRAINT, AND ADAPTATION IN THE VISUAL GENES OF NEOTROPICAL CICHLID FISHES AND OTHER VERTEBRATES Frances E. Hauser Doctor of Philosophy, 2018 Department of Ecology and Evolutionary Biology University of Toronto 2018 ABSTRACT The visual system serves as a direct interface between an organism and its environment. Studies of the molecular components of the visual transduction cascade, in particular visual pigments, offer an important window into the relationship between genetic variation and organismal fitness. In this thesis, I use molecular evolutionary models as well as protein modeling and experimental characterization to assess the role of variable evolutionary rates on visual protein function. In Chapter 2, I review recent work on the ecological and evolutionary forces giving rise to the impressive variety of adaptations found in visual pigments. In Chapter 3, I use interspecific vertebrate and mammalian datasets of two visual genes (RH1 or rhodopsin, and RPE65, a retinoid isomerase) to assess different methods for estimating evolutionary rate across proteins and the reliability of inferring evolutionary conservation at individual amino acid sites, with a particular emphasis on sites implicated in impaired protein function. ii In Chapters 4, and 5, I narrow my focus to devote particular attention to visual pigments in Neotropical cichlids, a highly diverse clade of fishes distributed across South and Central America. -

Summary Report of Freshwater Nonindigenous Aquatic Species in U.S
Summary Report of Freshwater Nonindigenous Aquatic Species in U.S. Fish and Wildlife Service Region 4—An Update April 2013 Prepared by: Pam L. Fuller, Amy J. Benson, and Matthew J. Cannister U.S. Geological Survey Southeast Ecological Science Center Gainesville, Florida Prepared for: U.S. Fish and Wildlife Service Southeast Region Atlanta, Georgia Cover Photos: Silver Carp, Hypophthalmichthys molitrix – Auburn University Giant Applesnail, Pomacea maculata – David Knott Straightedge Crayfish, Procambarus hayi – U.S. Forest Service i Table of Contents Table of Contents ...................................................................................................................................... ii List of Figures ............................................................................................................................................ v List of Tables ............................................................................................................................................ vi INTRODUCTION ............................................................................................................................................. 1 Overview of Region 4 Introductions Since 2000 ....................................................................................... 1 Format of Species Accounts ...................................................................................................................... 2 Explanation of Maps ................................................................................................................................ -

Neue Gattungseinteilung Der Mittelamerikanischen Cichliden
DCG_Info_07_2016_HR_20160621_DCG_Info 21.06.2016 06:51 Seite 146 Neue Gattungseinteilung der mittelamerikanischen Cichliden Rico Morgenstern Foto: Juan Miguel Artigas Azas Theraps irregularis verbleibt als einzige Art in der Gattung Theraps. Die Aufnahme entstand im Rio Lacanja im südlichen Chiapas, Mexiko. Inzwischen ist es fast 33 Jahre her, wenigstens Versuche, einzelne Gat- schien die Abhandlung „Diversity and dass KULLANDER (1983) die Gattung tungen neu zu definieren – aber eine evolution of the Middle American cich- Cichlasoma auf zwölf südamerikani- umfassende Gesamtbearbeitung er- lid fishes (Teleostei: Cichlidae) with re- sche, nahe mit der Typusart C. bima- folgte bisher nicht. Vielfach wurden vised classification“ (ŘIČAN et al. culatum verwandte Arten beschränkte. Zuordnungen vorgenommen, ohne 2016). Seither durfte der Name streng ge- dass man sich um eine wirkliche Be- nommen für die Mehrzahl der bis gründung bemühte. Die Arbeit berücksichtigt alle Cichliden dahin in dieser ehemaligen Sammel- Nord- und Mittelamerikas und der An- gattung untergebrachten, überwie- Bei der Gattungseinteilung der mittel- tillen sowie einige eng verwandte süd- gend mittelamerikanischen Arten amerikanischen Cichliden herrschte amerikanische Gattungen (Australoheros, nicht mehr verwendet werden. Man- somit bis vor kurzem ein ziemliches Caquetaia, Heroina, Mesoheros). gels geeigneter Alternativen ist das Chaos. Nun ist jedoch das Ende der An- Diese Fische gehören zu den „heroinen aber dennoch geschehen, wobei der führungszeichen (mit einer Ausnahme) -
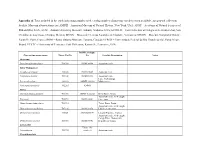
Appendix A. Taxa Included in the Study Indicating Samples Used, Catalog Number of Museum Vouchers When Available, and General Collection Locality
Appendix A. Taxa included in the study indicating samples used, catalog number of museum vouchers when available, and general collection locality. Museum abbreviations are: AMNH – American Museum of Natural History, New York, USA; ANSP – Academy of Natural Sciences of Philadelphia, USA; AUM – Auburn University Museum, Auburn, Alabama, USA; ECOSUR – Fish Collection at Colegio de la Frontera Sur, San Cristóbal de Las Casas, Chiapas, Mexico; MCNG – Museo de Ciencias Naturales de Guanare, Venezuela; MNHN – Muséum National d’Histoire Naturelle, Paris, France; ROM – Royal Ontario Museum, Toronto, Canada; UFRGS – Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil; UTFTC – University of Tennessee Fish Collection, Knoxville, Tennessee, USA. ROM Catalogue Current taxonomy name Tissue Cat No No Locality Description Notes Outgroup Pseudetroplus maculatus T14743 ROM 98998 Aquarium trade India-Madagascar Etroplus suratensis T13505 ROM 93809 Aquarium trade Paratilapia polleni T13100 ROM 88333 Aquarium trade Lake Andrapongy, Paretroplus damii 201936 AMNH 201936 Madagascar Paretroplus polyactis T12265 AMNH Africa Chromidotilapia guntheri T11700 AMNH I-226361 Beffa River, Benin Aquarium trade, wild caught, Etia nguti T10792 ROM 88042 Cameroon Hemichromis bimaculatus T11719 Tchan Duga, Benin Aquarium trade, wild caught, Heterochromis multidens T07136 ROM 88350 Lobeke, Cameroon Oreochromis niloticus 9092S AMNH254194 Littoral Province, Guinea Aquarium trade, wild caught, Congo River, Democratic Orthochromis stormsi T10766 ROM 88041 Republic of -

Cichlasoma Sieboldii and C. Tuba
Rev. Biol Trop., 23 (2) : 189-211, 1975 Taxonomy and biological aspects 01 the Central Amer'ican cichlid fishes Cichlasoma sieboldii and C. tuba by William A. Bussing'lí< (Received for publication May 22, 1975) ABSTRACTo' The complex taxonomic history of Cichlasoma sieboldii is re· viewed. Cichlasoma punctatum and its synonym Theraps terrabae are induded for the first time in the synoymy of C. sieboldii. A report of C. guttulatum from Pa namá is attributable to a misidentification of C. tuba, which had not been pre viously reported from that countIy. The morphologic and meristic variation, coloration, ecology and geographic distribution of C. sieboldii and C. tuba are discussed.. M:orphologic and distri butional evidence suggest that these twÜ' species are Pacific and Atlantic slope de rivatives of a common Pliocene' ancestor. There is a marked resemblance between the ciehlid fauna of the Atlan tic slope of lower Central Ameriea and the southeastern region of Costa Rica. The historieal reasons fo! this similarity have been discussed (4). The present study reviews the taxonomic history and discusses the biology of a Pacifie slope species, Ci¡h/asoma sieboldii, and its Atlantic versant counterpart, Cieh/asoma. tu ba (Fig. 1). MATERIAL AND METHODS Body measureinents are expressed in standard length (SL) in millime ters (mm). Body propartions of C. sieboldir as parts per mil appear in Table 1. The last two dorsal and anal rays are counted as ane only when their bases are in contact; when the base of the last ray is not touching the penultimate ray, each ray is counted separately. -

2011-Honduras-Fish-C
3 American Currents Vol. 41, No. 2 FROM THE “OL’ SOUTH” TO FARTHER SOUTH: KILLIES, MOLLIES, CICHLIDS, CHARACINS, AND KNIFEFISH IN HONDURAS Dustin Smith Lexington, South Carolina (with assistance from Rudy Arndt and Fritz Rohde) In March 2011, after a quick exploratory trip in 2008 by Fritz Rohde and Rudy Arndt, the three of us decided to make a return trip to Honduras. Fritz and Rudy went there in 2008 to visit a national park and potentially collect some killifish along the way. Fritz has an interest in Rivulus killies and we have (since then) spent significant amounts of time on trips to foreign lands chasing them. Doing an internet search on the fishes of Honduras, Fritz found that Wilfredo Matam- oros, a PhD graduate of the University of Southern Missis- sippi and currently (2011) a post-doc at LSU, was from Hon- duras and had published a paper on the freshwater fishes of his country (Matamoros et al. 2009). Will’s main interests were the Poeciliidae and their genetic relationships and he Figure 1. Honduras (http://tdhontario.tdh.ca). thought that there might be several undescribed species in Honduras. We made contact and Rudy decided to pay his 5 JAN 2011 travel expenses so he could join us on the trip and continue I arrived at the Atlanta airport early. As usual with every out his studies of Honduran fishes. We would assist with the col- of country expedition, I was excited and couldn’t sleep. Fritz lections (he had a permit), take photographs to be included arrived shortly afterwards and later so did Rudy. -

Exon-Based Phylogenomics Strengthens the Phylogeny of Neotropical Cichlids And
bioRxiv preprint doi: https://doi.org/10.1101/133512; this version posted July 13, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Exon-based phylogenomics strengthens the phylogeny of Neotropical cichlids and 2 identifies remaining conflicting clades (Cichliformes: Cichlidae: Cichlinae) 3 4 Katriina L. Ilves1,2*, Dax Torti3,4, and Hernán López-Fernández1 5 1 Department of Natural History, Royal Ontario Museum, 100 Queen’s Park, Toronto, 6 ON M5S 2C6 Canada 7 2 Current address: Biology Department, Pace University, 1 Pace Plaza, New York, NY 8 10038 9 3 Donnelly Sequencing Center, University of Toronto, 160 College Street, Toronto, ON 10 M5S 3E1 Canada 11 4 Current address: Ontario Institute for Cancer Research, MaRS Center, 661 University 12 Avenue, Toronto, ON M5G 0A3 Canada 13 *Corresponding author at: Biology Department, Pace University, 1 Pace Plaza, New 14 York, NY 10038; email: [email protected] 15 16 Abstract 17 The phenotypic, geographic, and species diversity of cichlid fishes have made them a 18 group of great interest for studying evolutionary processes. Here we present a targeted- 19 exon next-generation sequencing approach for investigating the evolutionary 20 relationships of cichlid fishes (Cichlidae), with focus on the Neotropical subfamily 21 Cichlinae using a set of 923 primarily single-copy exons designed through mining of the 22 Nile tilapia (Oreochromis niloticus) genome. Sequence capture and assembly were 23 robust, leading to a complete dataset of 415 exons for 139 species (147 terminals) that 1 bioRxiv preprint doi: https://doi.org/10.1101/133512; this version posted July 13, 2017. -

Teleostei: Cichlidae)
TERMS OF USE This pdf is provided by Magnolia Press for private/research use. Commercial sale or deposition in a public library or website is prohibited. Zootaxa 2833: 1–14 (2011) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2011 · Magnolia Press ISSN 1175-5334 (online edition) Paraneetroplus synspilus is a Junior Synonym of Paraneetroplus melanurus (Teleostei: Cichlidae) CALEB D. MCMAHAN1,2, CHRISTOPHER M. MURRAY2, AARON D. GEHEBER3,2, CHRISTOPHER D. BOECKMAN2 & KYLE R. PILLER2 1Division of Ichthyology, LSU Museum of Natural Science, Baton Rouge, LA 70803 USA. E-mail: [email protected] 2Department of Biological Sciences, Southeastern Louisiana University, Hammond LA 70402 USA 3Department of Zoology, University of Oklahoma, Norman OK 73019 USA Abstract The genus Paraneetroplus (Teleostei: Cichlidae) currently consists of 11 species that naturally occur from southern Mex- ico south to Panama. Paraneetroplus melanurus (Günther 1862) is found in the Lago de Petén system of Guatemala, and P. synspilus (Hubbs 1935) in the Río Grijalva-Usumacinta system, and other systems in Mexico, Belize, and Guatemala. Reported morphological differences between the two nominal species in the literature are vague but center around char- acteristics of a dark band that begins at the caudal fin and tapers anteriorly near mid-body. This band is reported as straight (horizontal) in P. melanurus but ventrally sloped in P. synspilus. Some authors have previously suggested that these two forms are not distinct. The purpose of this study was to conduct a systematic morphological comparison of P. melanurus and P. synspilus to further investigate their validity. We examined meristic, morphometric, and geometric morphometric characters and failed to recover diagnostic differences between these two forms. -

Paraneetroplus Synspilus Is a Junior Synonym of Paraneetroplus Melanurus (Teleostei: Cichlidae)
Zootaxa 2833: 1–14 (2011) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2011 · Magnolia Press ISSN 1175-5334 (online edition) Paraneetroplus synspilus is a Junior Synonym of Paraneetroplus melanurus (Teleostei: Cichlidae) CALEB D. MCMAHAN1,2, CHRISTOPHER M. MURRAY2, AARON D. GEHEBER3,2, CHRISTOPHER D. BOECKMAN2 & KYLE R. PILLER2 1Division of Ichthyology, LSU Museum of Natural Science, Baton Rouge, LA 70803 USA. E-mail: [email protected] 2Department of Biological Sciences, Southeastern Louisiana University, Hammond LA 70402 USA 3Department of Zoology, University of Oklahoma, Norman OK 73019 USA Abstract The genus Paraneetroplus (Teleostei: Cichlidae) currently consists of 11 species that naturally occur from southern Mex- ico south to Panama. Paraneetroplus melanurus (Günther 1862) is found in the Lago de Petén system of Guatemala, and P. synspilus (Hubbs 1935) in the Río Grijalva-Usumacinta system, and other systems in Mexico, Belize, and Guatemala. Reported morphological differences between the two nominal species in the literature are vague but center around char- acteristics of a dark band that begins at the caudal fin and tapers anteriorly near mid-body. This band is reported as straight (horizontal) in P. melanurus but ventrally sloped in P. synspilus. Some authors have previously suggested that these two forms are not distinct. The purpose of this study was to conduct a systematic morphological comparison of P. melanurus and P. synspilus to further investigate their validity. We examined meristic, morphometric, and geometric morphometric characters and failed to recover diagnostic differences between these two forms. The characters proposed to separate them do not allow for their differentiation, and we conclude that P. -

A New Species of Polymorphic Fish, Cichlasoma Minckleyi, from Cuatro Cienegas, Mexico (Teleostei: Cichlidae)
PROC. BIOL. SOC. WASH. 96(2), 1983, pp. 253-269 A NEW SPECIES OF POLYMORPHIC FISH, CICHLASOMA MINCKLEYI, FROM CUATRO CIENEGAS, MEXICO (TELEOSTEI: CICHLIDAE) Iry Kornfield and Jeffrey N. Taylor Abstract.—Cichlasorna rninckleyi is described from the Cuatro Cienegas basin, Coahuila, Mexico. Discrete morphological variants occurring sympatrically with- in this taxon incorporate differences normally separating distinct congeners. Vari- ation is partitioned into two non-overlapping body forms (deep-bodied and slen- der-bodied) within which occur two distinct pharyngeal morphs (papilliform and molariform), each maintained at high frequencies in natural populations. Field observations of matings both between morphs and between forms have estab- lished conspecificity of morphological variants. The dichotomous intraspecific variation presented by C. minckleyi suggests that phenetic characterization alone may be insufficient to delineate biological species within some members of the Cichlidae. An unusual neotropical cichlid has received considerable attention from evo- lutionary biologists because of discrete, pronounced morphological and trophic variation. The specific status of this fish had been unresolved because the mag- nitude of morphological variation suggested the existence of at least three closely related taxa (LaBounty 1974), while biochemical analysis strongly supported the existence of a single polymorphic species (Sage and Selander 1975). Recent field studies (Kornfield et al. 1982) have confirmed the biochemical analyses and es- tablished conspecificity among morphological variants. Described here is Cich- lasoma minckleyi, a new species of polymorphic cichlid endemic to the Cuatro Cienegas basin, Coahuila, Mexico. Methods Standard counts and measurements were recorded following the procedures of Taylor and Miller (1980). Measurements and qualitative shape descriptions of pharyngeal jaws were taken from Barel et al. -
Multilocus Phylogeny and Rapid Radiations in Neotropical Cichlid Fishes
Molecular Phylogenetics and Evolution 55 (2010) 1070–1086 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae) Hernán López-Fernández a,b,*, Kirk O. Winemiller c, Rodney L. Honeycutt d a Department of Natural History, Royal Ontario Museum, 100 Queen’s Park, Toronto, Ontario, Canada M5S 2C6 b Department of Ecology and Evolutionary Biology, University of Toronto, 25 Willcocks Street, Toronto, Ontario, Canada M5S 3B2 c Section of Ecology, Evolution and Systematic Biology, Department of Wildlife and Fisheries Sciences, Texas A&M University, College Station, TX 77843-2258, USA d Natural Science Division, Pepperdine University, 24255 Pacific Coast Hwy., Malibu, CA 90263, USA article info abstract Article history: Neotropical cichlid fishes comprise approximately 60 genera and at least 600 species, but despite this Received 14 September 2009 diversity, their phylogeny is only partially understood, which limits taxonomic, ecological and evolution- Revised 11 February 2010 ary research. We report the largest molecular phylogeny of Neotropical cichlids produced to date, com- Accepted 16 February 2010 bining data from three mitochondrial and two nuclear markers for 57 named genera and 154 species Available online 21 February 2010 from South and Central America. Neotropical cichlids (subfamily Cichlinae) were strongly monophyletic and were grouped into two main clades in -

Trophic Ecomorphology of Cichlid Fishes of Selva Lacandona, Usumacinta, Mexico
Environ Biol Fish https://doi.org/10.1007/s10641-019-00884-5 Trophic ecomorphology of cichlid fishes of Selva Lacandona, Usumacinta, Mexico Miriam Soria-Barreto & Rocío Rodiles-Hernández & Kirk O. Winemiller Received: 1 October 2018 /Accepted: 14 May 2019 # Springer Nature B.V. 2019 Abstract Neotropical cichlids exhibit great diversi- invertebrates and/or fish), with detritivores repre- ty of morphological traits associated with feeding, sented by relatively few species and strict piscivore by locomotion, and habitat use. We examined the one species. Dietary overlap was highest among carni- relationship between functional traits and diet by vores (P. friedrichsthalii and T. salvini), herbivores analyzing a dataset for 14 cichlid species from rivers in (C. intermedium and C. pearsei) and detritivore- the Selva Lacandona region, Usumacinta Basin, Chia- herbivores (V. melanura and K. ufermanni). Dietary pas, Mexico. Volumetric proportions of ingested food components were strongly correlated with several mor- items were used to calculate diet breath and interspecific phological traits, confirming patterns observed in other dietary overlap. Morphometric analysis was performed cichlids. For example, jaw protrusion and mandible using 24 traits associated with feeding. Associations length were positively correlated with consumption of between morphological and dietary components were fish and terrestrial invertebrates. A longer gut and a assessed using canonical correspondence analysis. wider tooth plate on the lower pharyngeal jaw were The most