Incidence Density Sampling for Nested Case-Control Study Designs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FDA Oncology Experience in Innovative Adaptive Trial Designs
Innovative Adaptive Trial Designs Rajeshwari Sridhara, Ph.D. Director, Division of Biometrics V Office of Biostatistics, CDER, FDA 9/3/2015 Sridhara - Ovarian cancer workshop 1 Fixed Sample Designs • Patient population, disease assessments, treatment, sample size, hypothesis to be tested, primary outcome measure - all fixed • No change in the design features during the study Adaptive Designs • A study that includes a prospectively planned opportunity for modification of one or more specified aspects of the study design and hypotheses based on analysis of data (interim data) from subjects in the study 9/3/2015 Sridhara - Ovarian cancer workshop 2 Bayesian Designs • In the Bayesian paradigm, the parameter measuring treatment effect is regarded as a random variable • Bayesian inference is based on the posterior distribution (Bayes’ Rule – updated based on observed data) – Outcome adaptive • By definition adaptive design 9/3/2015 Sridhara - Ovarian cancer workshop 3 Adaptive Designs (Frequentist or Bayesian) • Allows for planned design modifications • Modifications based on data accrued in the trial up to the interim time • Unblinded or blinded interim results • Control probability of false positive rate for multiple options • Control operational bias • Assumes independent increments of information 9/3/2015 Sridhara - Ovarian cancer workshop 4 Enrichment Designs – Prognostic or Predictive • Untargeted or All comers design: – post-hoc enrichment, prospective-retrospective designs – Marker evaluation after randomization (example: KRAS in cetuximab -
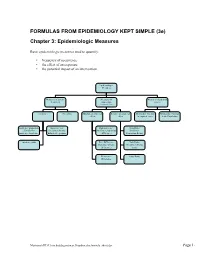
FORMULAS from EPIDEMIOLOGY KEPT SIMPLE (3E) Chapter 3: Epidemiologic Measures
FORMULAS FROM EPIDEMIOLOGY KEPT SIMPLE (3e) Chapter 3: Epidemiologic Measures Basic epidemiologic measures used to quantify: • frequency of occurrence • the effect of an exposure • the potential impact of an intervention. Epidemiologic Measures Measures of disease Measures of Measures of potential frequency association impact (“Measures of Effect”) Incidence Prevalence Absolute measures of Relative measures of Attributable Fraction Attributable Fraction effect effect in exposed cases in the Population Incidence proportion Incidence rate Risk difference Risk Ratio (Cumulative (incidence density, (Incidence proportion (Incidence Incidence, Incidence hazard rate, person- difference) Proportion Ratio) Risk) time rate) Incidence odds Rate Difference Rate Ratio (Incidence density (Incidence density difference) ratio) Prevalence Odds Ratio Difference Macintosh HD:Users:buddygerstman:Dropbox:eks:formula_sheet.doc Page 1 of 7 3.1 Measures of Disease Frequency No. of onsets Incidence Proportion = No. at risk at beginning of follow-up • Also called risk, average risk, and cumulative incidence. • Can be measured in cohorts (closed populations) only. • Requires follow-up of individuals. No. of onsets Incidence Rate = ∑person-time • Also called incidence density and average hazard. • When disease is rare (incidence proportion < 5%), incidence rate ≈ incidence proportion. • In cohorts (closed populations), it is best to sum individual person-time longitudinally. It can also be estimated as Σperson-time ≈ (average population size) × (duration of follow-up). Actuarial adjustments may be needed when the disease outcome is not rare. • In an open populations, Σperson-time ≈ (average population size) × (duration of follow-up). Examples of incidence rates in open populations include: births Crude birth rate (per m) = × m mid-year population size deaths Crude mortality rate (per m) = × m mid-year population size deaths < 1 year of age Infant mortality rate (per m) = × m live births No. -

Incidence and Secondary Transmission of SARS-Cov-2 Infections in Schools
Prepublication Release Incidence and Secondary Transmission of SARS-CoV-2 Infections in Schools Kanecia O. Zimmerman, MD; Ibukunoluwa C. Akinboyo, MD; M. Alan Brookhart, PhD; Angelique E. Boutzoukas, MD; Kathleen McGann, MD; Michael J. Smith, MD, MSCE; Gabriela Maradiaga Panayotti, MD; Sarah C. Armstrong, MD; Helen Bristow, MPH; Donna Parker, MPH; Sabrina Zadrozny, PhD; David J. Weber, MD, MPH; Daniel K. Benjamin, Jr., MD, PhD; for The ABC Science Collaborative DOI: 10.1542/peds.2020-048090 Journal: Pediatrics Article Type: Regular Article Citation: Zimmerman KO, Akinboyo IC, Brookhart A, et al. Incidence and secondary transmission of SARS-CoV-2 infections in schools. Pediatrics. 2021; doi: 10.1542/peds.2020- 048090 This is a prepublication version of an article that has undergone peer review and been accepted for publication but is not the final version of record. This paper may be cited using the DOI and date of access. This paper may contain information that has errors in facts, figures, and statements, and will be corrected in the final published version. The journal is providing an early version of this article to expedite access to this information. The American Academy of Pediatrics, the editors, and authors are not responsible for inaccurate information and data described in this version. Downloaded from©2021 www.aappublications.org/news American Academy by of guest Pediatrics on September 27, 2021 Prepublication Release Incidence and Secondary Transmission of SARS-CoV-2 Infections in Schools Kanecia O. Zimmerman, MD1,2,3; Ibukunoluwa C. Akinboyo, MD1,2; M. Alan Brookhart, PhD4; Angelique E. Boutzoukas, MD1,2; Kathleen McGann, MD2; Michael J. -

Estimated HIV Incidence and Prevalence in the United States
Volume 26, Number 1 Estimated HIV Incidence and Prevalence in the United States, 2015–2019 This issue of the HIV Surveillance Supplemental Report is published by the Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention (CDC), U.S. Department of Health and Human Services, Atlanta, Georgia. Estimates are presented for the incidence and prevalence of HIV infection among adults and adolescents (aged 13 years and older) based on data reported to CDC through December 2020. The HIV Surveillance Supplemental Report is not copyrighted and may be used and reproduced without permission. Citation of the source is, however, appreciated. Suggested citation Centers for Disease Control and Prevention. Estimated HIV incidence and prevalence in the United States, 2015–2019. HIV Surveillance Supplemental Report 2021;26(No. 1). http://www.cdc.gov/ hiv/library/reports/hiv-surveillance.html. Published May 2021. Accessed [date]. On the Web: http://www.cdc.gov/hiv/library/reports/hiv-surveillance.html Confidential information, referrals, and educational material on HIV infection CDC-INFO 1-800-232-4636 (in English, en Español) 1-888-232-6348 (TTY) http://wwwn.cdc.gov/dcs/ContactUs/Form Acknowledgments Publication of this report was made possible by the contributions of the state and territorial health departments and the HIV surveillance programs that provided surveillance data to CDC. This report was prepared by the following staff and contractors of the Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, CDC: Laurie Linley, Anna Satcher Johnson, Ruiguang Song, Sherry Hu, Baohua Wu, H. -

Understanding Relative Risk, Odds Ratio, and Related Terms: As Simple As It Can Get Chittaranjan Andrade, MD
Understanding Relative Risk, Odds Ratio, and Related Terms: As Simple as It Can Get Chittaranjan Andrade, MD Each month in his online Introduction column, Dr Andrade Many research papers present findings as odds ratios (ORs) and considers theoretical and relative risks (RRs) as measures of effect size for categorical outcomes. practical ideas in clinical Whereas these and related terms have been well explained in many psychopharmacology articles,1–5 this article presents a version, with examples, that is meant with a view to update the knowledge and skills to be both simple and practical. Readers may note that the explanations of medical practitioners and examples provided apply mostly to randomized controlled trials who treat patients with (RCTs), cohort studies, and case-control studies. Nevertheless, similar psychiatric conditions. principles operate when these concepts are applied in epidemiologic Department of Psychopharmacology, National Institute research. Whereas the terms may be applied slightly differently in of Mental Health and Neurosciences, Bangalore, India different explanatory texts, the general principles are the same. ([email protected]). ABSTRACT Clinical Situation Risk, and related measures of effect size (for Consider a hypothetical RCT in which 76 depressed patients were categorical outcomes) such as relative risks and randomly assigned to receive either venlafaxine (n = 40) or placebo odds ratios, are frequently presented in research (n = 36) for 8 weeks. During the trial, new-onset sexual dysfunction articles. Not all readers know how these statistics was identified in 8 patients treated with venlafaxine and in 3 patients are derived and interpreted, nor are all readers treated with placebo. These results are presented in Table 1. -

Disease Incidence, Prevalence and Disability
Part 3 Disease incidence, prevalence and disability 9. How many people become sick each year? 28 10. Cancer incidence by site and region 29 11. How many people are sick at any given time? 31 12. Prevalence of moderate and severe disability 31 13. Leading causes of years lost due to disability in 2004 36 World Health Organization 9. How many people become sick each such as diarrhoeal disease or malaria, it is common year? for individuals to be infected repeatedly and have several episodes. For such conditions, the number The “incidence” of a condition is the number of new given in the table is the number of disease episodes, cases in a period of time – usually one year (Table 5). rather than the number of individuals affected. For most conditions in this table, the figure given is It is important to remember that the incidence of the number of individuals who developed the illness a disease or condition measures how many people or problem in 2004. However, for some conditions, are affected by it for the first time over a period of Table 5: Incidence (millions) of selected conditions by WHO region, 2004 Eastern The Mediter- South- Western World Africa Americas ranean Europe East Asia Pacific Tuberculosisa 7.8 1.4 0.4 0.6 0.6 2.8 2.1 HIV infectiona 2.8 1.9 0.2 0.1 0.2 0.2 0.1 Diarrhoeal diseaseb 4 620.4 912.9 543.1 424.9 207.1 1 276.5 1 255.9 Pertussisb 18.4 5.2 1.2 1.6 0.7 7.5 2.1 Measlesa 27.1 5.3 0.0e 1.0 0.2 17.4 3.3 Tetanusa 0.3 0.1 0.0 0.1 0.0 0.1 0.0 Meningitisb 0.7 0.3 0.1 0.1 0.0 0.2 0.1 Malariab 241.3 203.9 2.9 8.6 0.0 23.3 2.7 -

Incidence, Risk, and Case Fatality of First Ever Stroke in the Elderly Population
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.74.3.317 on 1 March 2003. Downloaded from 317 PAPER Incidence, risk, and case fatality of first ever stroke in the elderly population. The Rotterdam Study M Hollander, P J Koudstaal, M L Bots, D E Grobbee, A Hofman,MMBBreteler ............................................................................................................................. J Neurol Neurosurg Psychiatry 2003;74:317–321 See end of article for authors’ affiliations ....................... Objective: To estimate the incidence, survival, and lifetime risk of stroke in the elderly population. Methods: The authors conducted a study in 7721 participants from the population based Rotterdam Correspondence to: Dr M M.B. Breteler, Study who were free from stroke at baseline (1990–1993) and were followed up for stroke until Department of 1 January 1999. Age and sex specific incidence, case fatality rates, and lifetime risks of stroke were Epidemiology and calculated. Biostatistics, Erasmus Results: Mean follow up was 6.0 years and 432 strokes occurred. The incidence rate of stroke per Medical Centre Rotterdam, 1000 person years increased with age and ranged from 1.7 (95% CI 0.4 to 6.6) in men aged 55 to PO Box 1738, 3000 DR Rotterdam, Netherlands; 59 years to 69.8 (95% CI 22.5 to 216.6) in men aged 95 years or over. Corresponding figures for [email protected] women were 1.2 (95% CI 0.3 to 4.7) and 33.1 (95% CI 17.8 to 61.6). Men and women had similar absolute lifetime risks of stroke (21% for those aged 55 years). The survival after stroke did not differ Received 17 April 2002 Accepted in revised form according to sex. -

EPI Case Study 1 Incidence, Prevalence, and Disease
EPIDEMIOLOGY CASE STUDY 1: Incidence, Prevalence, and Disease Surveillance; Historical Trends in the Epidemiology of M. tuberculosis STUDENT VERSION 1.0 EPI Case Study 1: Incidence, Prevalence, and Disease Surveillance; Historical Trends in the Epidemiology of M. tuberculosis Estimated Time to Complete Exercise: 30 minutes LEARNING OBJECTIVES At the completion of this Case Study, participants should be able to: ¾ Explain why denominators are necessary when comparing changes in morbidity and mortality over time ¾ Distinguish between incidence rates and prevalence ratios ¾ Calculate and interpret cause-specific morbidity and mortality rates ¾ Describe how changes in mortality or morbidity could be due to an artifact rather than a real change ASPH EPIDEMIOLOGY COMPETENCIES ADDRESSED C. 3. Describe a public health problem in terms of magnitude, person, place, and time C. 6. Apply the basic terminology and definitions of epidemiology C. 7. Calculate basic epidemiology measures C. 9. Draw appropriate inference from epidemiologic data C. 10. Evaluate the strengths and limitations of epidemiologic reports ASPH INTERDISCIPLINARY/CROSS-CUTTING COMPETENCIES ADDRESSED F.1. [Communication and Informatics] Describe how the public health information infrastructure is used to collect, process, maintain, and disseminate data J.1. [Professionalism] Discuss sentinel events in the history and development of the public health profession and their relevance for practice in the field L.2. [Systems Thinking] Identify unintended consequences produced by changes made to a public health system This material was developed by the staff at the Global Tuberculosis Institute (GTBI), one of four Regional Training and Medical Consultation Centers funded by the Centers for Disease Control and Prevention. It is published for learning purposes only. -

Epidemiologic Study Designs
Epidemiologic Study Designs Jacky M Jennings, PhD, MPH Associate Professor Associate Director, General Pediatrics and Adolescent Medicine Director, Center for Child & Community Health Research (CCHR) Departments of Pediatrics & Epidemiology Johns Hopkins University Learning Objectives • Identify basic epidemiologic study designs and their frequent sequence of study • Recognize the basic components • Understand the advantages and disadvantages • Appropriately select a study design Research Question & Hypotheses Analytic Study Plan Design Basic Study Designs and their Hierarchy Clinical Observation Hypothesis Descriptive Study Case-Control Study Cohort Study Randomized Controlled Trial Systematic Review Causality Adapted from Gordis, 1996 MMWR Study Design in Epidemiology • Depends on: – The research question and hypotheses – Resources and time available for the study – Type of outcome of interest – Type of exposure of interest – Ethics Study Design in Epidemiology • Includes: – The research question and hypotheses – Measures and data quality – Time – Study population • Inclusion/exclusion criteria • Internal/external validity Epidemiologic Study Designs • Descriptive studies – Seeks to measure the frequency of disease and/or collect descriptive data on risk factors • Analytic studies – Tests a causal hypothesis about the etiology of disease • Experimental studies – Compares, for example, treatments EXPOSURE Cross-sectional OUTCOME EXPOSURE OUTCOME Case-Control EXPOSURE OUTCOME Cohort TIME Cross-sectional studies • Measure existing disease and current exposure levels at one point in time • Sample without knowledge of exposure or disease • Ex. Prevalence studies Cross-sectional studies • Advantages – Often early study design in a line of investigation – Good for hypothesis generation – Relatively easy, quick and inexpensive…depends on question – Examine multiple exposures or outcomes – Estimate prevalence of disease and exposures Cross-sectional studies • Disadvantages – Cannot infer causality – Prevalent vs. -

Incidence of Drug Injection
Incidence of drug injection: systematic review and meta-analysis of cohort studies among at risk populations María J Bravo Blanca I Indave EMCDDA annual expert meeting on Drug-related deaths (DRD) & Drug-related infectious diseases (DRID) 16-18 October 2013 – EMCDDA (Lisbon) Background • Initiation into drug injection is an important determinant of morbidity and mortality (blood-borne infections …HIV, HCV … and overdose). • Incidence of drug injection (IDI) is relevat Prevention/projections • IDI Cohort studies never injectors of illegal drugs/vulnerable lifestyle • Cohort studies (Coh-S) – potential of bias – Validity (internal/external) – Coh-S of hidden population presents difficultis for recruitement + high attrition lack of statistical power • There is remarkable heterogeneity between theIDI of well-known cohorts • No systematic reviews published AIMS 1. To carry out a Systematic Review of cohort studies that estimate IDI among never drug injectors at risk 2. To conduct a meta-analysis pooled IDI and explore sources of heterogeneity and bias Methods -Systematic review • Search for Cohort Studies on initiation into DI among vulnerable pop • EMBASE, Lilacs, Medline, PsycINFO. Cochrane database. 1980-2012 • MeSH, key words. No language restrictions. Published/grey literature. • Data extraction: Two independent reviewers. STROBE guidelines. • Standardize quality assessment form (SIGN50 –Scottish Intercollegiate Network-) & Drug related check list (NDARC). • Inclusion criteria: Cohort studies on initiation on drug injection: “the -

Epidemiology
5 Defining the current situation – epidemiology Paul R Hunter and Helen Risebro The first step in any economic appraisal or evaluation is to understand the underlying problem being addressed (see Chapter 1). Clearly, such an analysis of drinking-water interventions will have a strong public health element. This chapter discusses the role of epidemiology in identifying the burden of disease1 in a community that may be attributable to lack of access to safe drinking-water or adequate sanitation. In order to determine the scale of the problem, there are three questions to be asked: • What is the burden of disease in the target group? 1 WHO measures the burden of disease in disability-adjusted life years (DALYs). This time-bound measurement combines years of life lost as a result of premature death and years of healthy life lost because of time lived in a status of less than full health. Mortality and morbidity are linked to other indicators such as financial costs. © 2011 World Health Organization (WHO). Valuing Water, Valuing Livelihoods. Edited by John Cameron, Paul Hunter, Paul Jagals and Katherine Pond. Published by IWA Publishing, London, UK. 76 Valuing Water, Valuing Livelihoods • What proportion of the burden of disease is caused by deficiencies in access to drinking-water that are to be remedied by the intervention? • Are there any spin-off livelihood effects that would result from the outcomes of the intervention? This chapter focuses on the first two questions. Specific data challenges related to livelihood analysis are raised in Chapter 6. This chapter aims to assist decision-makers in gathering evidence to enable them to make an informed decision about whether or not there is a public health need for an intervention. -

FDA Briefing Document: Pfizer-Biontech COVID-19 Vaccine
Vaccines and Related Biological Products Advisory Committee Meeting December 10, 2020 FDA Briefing Document Pfizer-BioNTech COVID-19 Vaccine Sponsor: Pfizer and BioNTech Table of Contents List of Tables ............................................................................................................................. 3 List of Figures ............................................................................................................................ 4 Glossary..................................................................................................................................... 5 1. Executive Summary ............................................................................................................... 6 2. Background ............................................................................................................................ 7 2.1. SARS-CoV-2 Pandemic ................................................................................................ 7 2.2. EUA Request for the Pfizer and BioNTech COVID-19 Vaccine BNT162b2 .................... 8 2.3. U.S. Requirements to Support Issuance of an EUA for a Biological Product ........................................................................................................................... 8 2.4. Applicable Guidance for Industry ................................................................................... 9 2.5. Safety and Effectiveness Information Needed to Support an EUA ................................. 9 2.6. Continuation