U. C. Banerjee, Ph.D. 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall Et Al
USOO9689046B2 (12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall et al. (45) Date of Patent: Jun. 27, 2017 (54) SYSTEM AND METHODS FOR THE FOREIGN PATENT DOCUMENTS DETECTION OF MULTIPLE CHEMICAL WO O125472 A1 4/2001 COMPOUNDS WO O169245 A2 9, 2001 (71) Applicants: Robert Matthew Mayall, Calgary (CA); Emily Candice Hicks, Calgary OTHER PUBLICATIONS (CA); Margaret Mary-Flora Bebeselea, A. et al., “Electrochemical Degradation and Determina Renaud-Young, Calgary (CA); David tion of 4-Nitrophenol Using Multiple Pulsed Amperometry at Christopher Lloyd, Calgary (CA); Lisa Graphite Based Electrodes', Chem. Bull. “Politehnica” Univ. Kara Oberding, Calgary (CA); Iain (Timisoara), vol. 53(67), 1-2, 2008. Fraser Scotney George, Calgary (CA) Ben-Yoav. H. et al., “A whole cell electrochemical biosensor for water genotoxicity bio-detection”. Electrochimica Acta, 2009, 54(25), 6113-6118. (72) Inventors: Robert Matthew Mayall, Calgary Biran, I. et al., “On-line monitoring of gene expression'. Microbi (CA); Emily Candice Hicks, Calgary ology (Reading, England), 1999, 145 (Pt 8), 2129-2133. (CA); Margaret Mary-Flora Da Silva, P.S. et al., “Electrochemical Behavior of Hydroquinone Renaud-Young, Calgary (CA); David and Catechol at a Silsesquioxane-Modified Carbon Paste Elec trode'. J. Braz. Chem. Soc., vol. 24, No. 4, 695-699, 2013. Christopher Lloyd, Calgary (CA); Lisa Enache, T. A. & Oliveira-Brett, A. M., "Phenol and Para-Substituted Kara Oberding, Calgary (CA); Iain Phenols Electrochemical Oxidation Pathways”, Journal of Fraser Scotney George, Calgary (CA) Electroanalytical Chemistry, 2011, 1-35. Etesami, M. et al., “Electrooxidation of hydroquinone on simply prepared Au-Pt bimetallic nanoparticles'. Science China, Chem (73) Assignee: FREDSENSE TECHNOLOGIES istry, vol. -

Bioactive Principles in Two Polyherbal Traditional Anti- Diabeteic Formulations
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN:2278-3008, p-ISSN:2319-7676. Volume 12, Issue 5 Ver. III (Sep. – Oct. 2017), PP 75-83 www.iosrjournals.org Bioactive Principles in Two Polyherbal Traditional Anti- Diabeteic Formulations *Ogbuji C A., 1 Ndulaka J.C. 2 Onuh, Emeka F 2 . and Eke Ugorji I. 3 1. Department of food Science and Technology, Abia State Polytechnic. Aba. Nigeria. 2. Department of Chemistry, Abia State Polytechnic, Aba. 3. Department of Medical Biochemistry, Abia State University, Uturu. Corresponding Author: Ogbuji C A., Abstract: Bioactive principles in two polyherbal traditional anti diabetic formulations of different plants used in the treatment of diabetes mellitus mixed in different ratios were characterized using Infrared spectroscopy. Six medicinal plants with proven anti diabetic and related beneficial effects were selected for the preparation of two mixtures Acanthus montanus, Asystasia gangetica, Emilia coccinea, Hibiscus rosa-sinesis, Momordica charantia (Bitter melon), and Venonia amygdalina. Mixtures of the All- Six (AS) herbal leaves recorded these compounds 3-beta-acetoxy-5-etienic, acid dihydroxyacetone, acetobromo-alpha-D-galactose, dihydroxyacetone, ethylacetohydroxomate, P-tolyacetonitrile, 4-aminoacetophenone, dihydroxyacetone,ethylacetohydroxomate, ethyl-4-chloro-2-cyanoacetoacetate while mixtures of All-four (AF) herbal leaves recorded ethylacetohydroxamate, ethyl-4,4,4-trichloroacetate, 4 – amino – acetophenone, ethylacetohydroxamate, p – tolyacetonitrile, thiophene-2-acetonitrile, ethyl-4-chloro-2-cyanoacetoacetate, acetobromo-alpha-D-galactose, ethylacetohydroxamate, 4-aminoacetophenone and thiophene-2-acetonitrile. Functional groups such as those of the nitriles, benzene, acetals, cyano-compounds, amines, amides, substituted and conjugated ketones and aldehydes, alkaloids, phenyl groups, chlorocompounds, bromo sugar, glycosides, thiophene derivatives, amino substituted compounds and indoles were identified. -

Comparison of Nitrile Hydratases in Rhodococcus Rhodochrous DAP 96253 and DAP 96622 Growing on Inducing and Non- Inducing Media
Georgia State University ScholarWorks @ Georgia State University Biology Dissertations Department of Biology Spring 4-26-2013 Comparison of Nitrile Hydratases in Rhodococcus Rhodochrous DAP 96253 and DAP 96622 Growing on Inducing and Non- Inducing Media Fengkun Du Georgia State University Follow this and additional works at: https://scholarworks.gsu.edu/biology_diss Recommended Citation Du, Fengkun, "Comparison of Nitrile Hydratases in Rhodococcus Rhodochrous DAP 96253 and DAP 96622 Growing on Inducing and Non-Inducing Media." Dissertation, Georgia State University, 2013. https://scholarworks.gsu.edu/biology_diss/130 This Dissertation is brought to you for free and open access by the Department of Biology at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Biology Dissertations by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. COMPARISON OF NITRILE HYDRATASES IN RHODOCOCCUS RHODOCHROUS DAP 96253 AND DAP 96622 GROWING ON INDUCING AND NON INDUCING MEDIA by FENGKUN DU Under the Direction of George E. Pierce ABSTRACT Nitrile hydratase activity in Rhodococcus rhodochrous DAP 96253 can be induced with multiple inducers that include urea, cobalt (Co), iron (Fe) and nickel (Ni). When induced with Co/urea, cells of R. rhodochrous DAP 96253 expressed the highest level of nitrile hydratase activity (~200 units/min·mg-cdw) when compared with the other inducers tested. Cells induced with Co had the second highest nitrile hydratase activity (~7 units/min·mg-cdw), whereas in the uninduced cells, nitrile hydratase activity was lower than 1 unit/min·mg-cdw. Similarly in R. rhodochrous DAP 96622, when induced with Co/urea, the nitrile hydratase activity of R. -

Analysis of the Sll0783 Function in PHB Synthesis in Synechocystis PCC 6803: a Crucial Role of NADPH
Analysis of the Sll0783 Function in PHB Synthesis in Synechocystis PCC 6803: a Crucial Role of NADPH in N-Starvation Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität Tübingen zur Erlangung des Grades eines Doktors der Naturwissenschaften (Dr. rer. nat.) vorgelegt von Maximilian Schlebusch aus Engelskirchen Tübingen 2012 Tag der mündlichen Qualifikation: 03.02.2012 Dekan: Prof. Dr. Wolfgang Rosenstiel 1. Berichterstatter: Prof. Dr. Karl Forchhammer 2. Berichterstatter: Prof. Dr. Wolfgang Wohlleben Abstract Nitrogen frequently is a limiting nutrient in natural habitats. Therefore, cyanobac- teria as well as other autotrophic organisms have developed multiple strategies to adapt to nitrogen deficiency. Transcriptomic analyses of the strain Synechocys- tis PCC 6803 under nitrogen-deficient conditions revealed a highly induced gene (sll0783 ), which is annotated as conserved protein with unknown function. This gene is part of a cluster with seven genes and in the upstream region lies a pre- dicted NtcA-binding site. Homologues of this cluster occur in some unicellular, non-diazotrophic cyanobacteria, in several α-, β- and γ-proteobacteria as well as in some gram-positives. The common link between the heterotrophic bacteria seems to be the ability of nitrogen fixation and production of polyhydroxybu- tyrate (PHB), whereas among the cyanobacteria only Synechocystis PCC 6803 can accumulate PHB. In this work, a knockout mutant of this gene in Synechocystis PCC 6803 was characterised. This mutant was unable to accumulate PHB, a carbon and en- ergy storage compound. The levels of precursor metabolites such as glycogen and acetyl-CoA were not reduced. The impairment in PHB accumulation cor- related with a loss of PHB synthase activity during prolonged nitrogen starva- tion. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

(12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur Et Al
US 2012026.6329A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur et al. (43) Pub. Date: Oct. 18, 2012 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/10 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/24 (2006.01) CI2N 9/02 (2006.01) (75) Inventors: Eric J. Mathur, Carlsbad, CA CI2N 9/06 (2006.01) (US); Cathy Chang, San Marcos, CI2P 2L/02 (2006.01) CA (US) CI2O I/04 (2006.01) CI2N 9/96 (2006.01) (73) Assignee: BP Corporation North America CI2N 5/82 (2006.01) Inc., Houston, TX (US) CI2N 15/53 (2006.01) CI2N IS/54 (2006.01) CI2N 15/57 2006.O1 (22) Filed: Feb. 20, 2012 CI2N IS/60 308: Related U.S. Application Data EN f :08: (62) Division of application No. 1 1/817,403, filed on May AOIH 5/00 (2006.01) 7, 2008, now Pat. No. 8,119,385, filed as application AOIH 5/10 (2006.01) No. PCT/US2006/007642 on Mar. 3, 2006. C07K I4/00 (2006.01) CI2N IS/II (2006.01) (60) Provisional application No. 60/658,984, filed on Mar. AOIH I/06 (2006.01) 4, 2005. CI2N 15/63 (2006.01) Publication Classification (52) U.S. Cl. ................... 800/293; 435/320.1; 435/252.3: 435/325; 435/254.11: 435/254.2:435/348; (51) Int. Cl. 435/419; 435/195; 435/196; 435/198: 435/233; CI2N 15/52 (2006.01) 435/201:435/232; 435/208; 435/227; 435/193; CI2N 15/85 (2006.01) 435/200; 435/189: 435/191: 435/69.1; 435/34; CI2N 5/86 (2006.01) 435/188:536/23.2; 435/468; 800/298; 800/320; CI2N 15/867 (2006.01) 800/317.2: 800/317.4: 800/320.3: 800/306; CI2N 5/864 (2006.01) 800/312 800/320.2: 800/317.3; 800/322; CI2N 5/8 (2006.01) 800/320.1; 530/350, 536/23.1: 800/278; 800/294 CI2N I/2 (2006.01) CI2N 5/10 (2006.01) (57) ABSTRACT CI2N L/15 (2006.01) CI2N I/19 (2006.01) The invention provides polypeptides, including enzymes, CI2N 9/14 (2006.01) structural proteins and binding proteins, polynucleotides CI2N 9/16 (2006.01) encoding these polypeptides, and methods of making and CI2N 9/20 (2006.01) using these polynucleotides and polypeptides. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

(12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur Et Al
US 20150240226A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur et al. (43) Pub. Date: Aug. 27, 2015 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/16 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/02 (2006.01) CI2N 9/78 (2006.01) (71) Applicant: BP Corporation North America Inc., CI2N 9/12 (2006.01) Naperville, IL (US) CI2N 9/24 (2006.01) CI2O 1/02 (2006.01) (72) Inventors: Eric J. Mathur, San Diego, CA (US); CI2N 9/42 (2006.01) Cathy Chang, San Marcos, CA (US) (52) U.S. Cl. CPC. CI2N 9/88 (2013.01); C12O 1/02 (2013.01); (21) Appl. No.: 14/630,006 CI2O I/04 (2013.01): CI2N 9/80 (2013.01); CI2N 9/241.1 (2013.01); C12N 9/0065 (22) Filed: Feb. 24, 2015 (2013.01); C12N 9/2437 (2013.01); C12N 9/14 Related U.S. Application Data (2013.01); C12N 9/16 (2013.01); C12N 9/0061 (2013.01); C12N 9/78 (2013.01); C12N 9/0071 (62) Division of application No. 13/400,365, filed on Feb. (2013.01); C12N 9/1241 (2013.01): CI2N 20, 2012, now Pat. No. 8,962,800, which is a division 9/2482 (2013.01); C07K 2/00 (2013.01); C12Y of application No. 1 1/817,403, filed on May 7, 2008, 305/01004 (2013.01); C12Y 1 1 1/01016 now Pat. No. 8,119,385, filed as application No. PCT/ (2013.01); C12Y302/01004 (2013.01); C12Y US2006/007642 on Mar. 3, 2006. -

Supplementary Material (ESI) for Natural Product Reports
Electronic Supplementary Material (ESI) for Natural Product Reports. This journal is © The Royal Society of Chemistry 2014 Supplement to the paper of Alexey A. Lagunin, Rajesh K. Goel, Dinesh Y. Gawande, Priynka Pahwa, Tatyana A. Gloriozova, Alexander V. Dmitriev, Sergey M. Ivanov, Anastassia V. Rudik, Varvara I. Konova, Pavel V. Pogodin, Dmitry S. Druzhilovsky and Vladimir V. Poroikov “Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review” Contents PASS (Prediction of Activity Spectra for Substances) Approach S-1 Table S1. The lists of 122 known therapeutic effects for 50 analyzed medicinal plants with accuracy of PASS prediction calculated by a leave-one-out cross-validation procedure during the training and number of active compounds in PASS training set S-6 Table S2. The lists of 3,345 mechanisms of action that were predicted by PASS and were used in this study with accuracy of PASS prediction calculated by a leave-one-out cross-validation procedure during the training and number of active compounds in PASS training set S-9 Table S3. Comparison of direct PASS prediction results of known effects for phytoconstituents of 50 TIM plants with prediction of known effects through “mechanism-effect” and “target-pathway- effect” relationships from PharmaExpert S-79 S-1 PASS (Prediction of Activity Spectra for Substances) Approach PASS provides simultaneous predictions of many types of biological activity (activity spectrum) based on the structure of drug-like compounds. The approach used in PASS is based on the suggestion that biological activity of any drug-like compound is a function of its structure. -

Springer Handbook of Enzymes
Dietmar Schomburg Ida Schomburg (Eds.) Springer Handbook of Enzymes Alphabetical Name Index 1 23 © Springer-Verlag Berlin Heidelberg New York 2010 This work is subject to copyright. All rights reserved, whether in whole or part of the material con- cerned, specifically the right of translation, printing and reprinting, reproduction and storage in data- bases. The publisher cannot assume any legal responsibility for given data. Commercial distribution is only permitted with the publishers written consent. Springer Handbook of Enzymes, Vols. 1–39 + Supplements 1–7, Name Index 2.4.1.60 abequosyltransferase, Vol. 31, p. 468 2.7.1.157 N-acetylgalactosamine kinase, Vol. S2, p. 268 4.2.3.18 abietadiene synthase, Vol. S7,p.276 3.1.6.12 N-acetylgalactosamine-4-sulfatase, Vol. 11, p. 300 1.14.13.93 (+)-abscisic acid 8’-hydroxylase, Vol. S1, p. 602 3.1.6.4 N-acetylgalactosamine-6-sulfatase, Vol. 11, p. 267 1.2.3.14 abscisic-aldehyde oxidase, Vol. S1, p. 176 3.2.1.49 a-N-acetylgalactosaminidase, Vol. 13,p.10 1.2.1.10 acetaldehyde dehydrogenase (acetylating), Vol. 20, 3.2.1.53 b-N-acetylgalactosaminidase, Vol. 13,p.91 p. 115 2.4.99.3 a-N-acetylgalactosaminide a-2,6-sialyltransferase, 3.5.1.63 4-acetamidobutyrate deacetylase, Vol. 14,p.528 Vol. 33,p.335 3.5.1.51 4-acetamidobutyryl-CoA deacetylase, Vol. 14, 2.4.1.147 acetylgalactosaminyl-O-glycosyl-glycoprotein b- p. 482 1,3-N-acetylglucosaminyltransferase, Vol. 32, 3.5.1.29 2-(acetamidomethylene)succinate hydrolase, p. 287 Vol. -

CYANIDE DEGRADATION by Bacillus Pumilus Cl: CELLULAR and MOLECULAR CHARACTERIZATION
\. CYANIDE DEGRADATION BY Bacillus pumilus Cl: CELLULAR AND MOLECULAR CHARACTERIZATION by Paul Robert Meyers A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy m the Department of Microbiology, Faculty of Science, University of Cape Town, South Africa. University of Cape Town Cape Town August 1993 The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town CERTIFICATION OF SUPERVISOR In termsof paragraph GP 8 of "General Rules for the degree of Doctor of Philosophy (PhD)" I, as supervisor of the candidate Paul R. Meyers, certify that I approve of the incorporationin this thesis of material that has already been published or submitted for publication. Pro essor D.R. Woods Deputy Vice Chancellor (Research) Uruversityof Cape Town Director of Microbial Research Units ACKNOWLEDGEMENTS I wish to record my grateful thanks to my supervisors Professor Dave Woods and Professor Doug Rawlings for supervising this research. Thank you for your enthusiasm and expert guidance in directing me to this goal. Special thanks are due to Dr George Lindsey of the Department of Biochemistry who was always available for discussion and who enthusiastically supervised a large section of this work. Thanks to Pravin Gokool of National Chemical Products for his assistance in establishing this project at the University of Cape Town and for his helpful advice and discussions. -
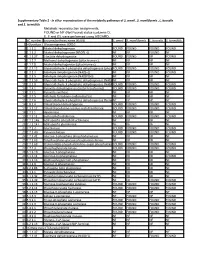
Supplementary Table 2 - in Silico Reconstruction of the Metabolic Pathways of S
Supplementary Table 2 - In silico reconstruction of the metabolic pathways of S. amnii , S. moniliformis , L. buccalis and S. termiditis Metabolic reconstruction assignments, FOUND or NF (Not Found) status (columns D, E, F and G), were performed using ASGARD, EC number Enzyme/pathway name (KEGG) S. amnii S. moniliformis L. buccalis S. termiditis 1 >Glycolysis / Gluconeogenesis 00010 2 1.1.1.1 Alcohol dehydrogenase. FOUND FOUND FOUND FOUND 3 1.1.1.2 Alcohol dehydrogenase (NADP(+)). NF NF FOUND NF 4 1.1.1.27 L-lactate dehydrogenase. FOUND FOUND NF FOUND 5 1.1.2.7 Methanol dehydrogenase (cytochrome c). NF NF NF NF 6 1.1.2.8 Alcohol dehydrogenase (cytochrome c). NF NF NF NF 7 1.2.1.12 Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating).FOUND FOUND FOUND FOUND 8 1.2.1.3 Aldehyde dehydrogenase (NAD(+)). NF NF FOUND FOUND 9 1.2.1.5 Aldehyde dehydrogenase (NAD(P)(+)). NF NF NF NF 10 1.2.1.59 Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+))NF (phosphorylating).NF NF NF 11 1.2.1.9 Glyceraldehyde-3-phosphate dehydrogenase (NADP(+)).FOUND FOUND FOUND FOUND 12 1.2.4.1 Pyruvate dehydrogenase (acetyl-transferring). FOUND FOUND FOUND FOUND 13 1.2.7.1 Pyruvate synthase. NF NF NF NF 14 1.2.7.5 Aldehyde ferredoxin oxidoreductase. NF NF NF NF 15 1.2.7.6 Glyceraldehyde-3-phosphate dehydrogenase (ferredoxin).NF NF NF NF 16 1.8.1.4 Dihydrolipoyl dehydrogenase. FOUND FOUND FOUND FOUND 17 2.3.1.12 Dihydrolipoyllysine-residue acetyltransferase. FOUND FOUND FOUND FOUND 18 2.7.1.1 Hexokinase.