Springer Handbook of Enzymes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Review Article Functional Subunits of Eukaryotic Chaperonin CCT/Tric in Protein Folding
SAGE-Hindawi Access to Research Journal of Amino Acids Volume 2011, Article ID 843206, 16 pages doi:10.4061/2011/843206 Review Article Functional Subunits of Eukaryotic Chaperonin CCT/TRiC in Protein Folding M. Anaul Kabir,1 Wasim Uddin,1 Aswathy Narayanan,1 Praveen Kumar Reddy,1 M. Aman Jairajpuri,2 Fred Sherman,3 and Zulfiqar Ahmad4 1 Molecular Genetics Laboratory, School of Biotechnology, National Institute of Technology Calicut, Kerala 673601, India 2 Department of Biosciences, Jamia Millia Islamia, Jamia Nagar, New Delhi 110025, India 3 Department of Biochemistry and Biophysics, University of Rochester Medical Center, NY 14642, USA 4 Department of Biology, Alabama A&M University, Normal, AL 35762, USA Correspondence should be addressed to M. Anaul Kabir, [email protected] Received 15 February 2011; Accepted 5 April 2011 Academic Editor: Shandar Ahmad Copyright © 2011 M. Anaul Kabir et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Molecular chaperones are a class of proteins responsible for proper folding of a large number of polypeptides in both prokaryotic and eukaryotic cells. Newly synthesized polypeptides are prone to nonspecific interactions, and many of them make toxic aggregates in absence of chaperones. The eukaryotic chaperonin CCT is a large, multisubunit, cylindrical structure having two identical rings stacked back to back. Each ring is composed of eight different but similar subunits and each subunit has three distinct domains. CCT assists folding of actin, tubulin, and numerous other cellular proteins in an ATP-dependent manner. -

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

Structural Insights Into Histone Modifying Enzymes
Wayne State University Wayne State University Theses January 2019 Structural Insights Into Histone Modifying Enzymes Shruti Amle Wayne State University, [email protected] Follow this and additional works at: https://digitalcommons.wayne.edu/oa_theses Part of the Biochemistry Commons, and the Molecular Biology Commons Recommended Citation Amle, Shruti, "Structural Insights Into Histone Modifying Enzymes" (2019). Wayne State University Theses. 693. https://digitalcommons.wayne.edu/oa_theses/693 This Open Access Embargo is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Theses by an authorized administrator of DigitalCommons@WayneState. STRUCTURAL INSIGHTS INTO HISTONE MODIFYING ENZYMES by SHRUTI AMLE THESIS Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE 2019 MAJOR: BIOCHEMISTRY AND MOLECULAR BIOLOGY Approved By: _________________________________________ Advisor Date _________________________________________ _________________________________________ _________________________________________ i ACKNOWLEDGEMENTS Writing this thesis has been extremely captivating and gratifying. I take this opportunity to express my deep and sincere acknowledgements to the number of people for extending their generous support and unstinted help during my entire study. Firstly, I would like to express my respectful regards and deep sense of gratitude to my advisor, Dr. Zhe Yang. I am extremely honored to study and work under his guidance. His vision, ideals, timely motivation and immense knowledge had a deep influence on my entire journey of this career. Without his understanding and support, it would not have been possible to complete this research successfully. I also owe my special thanks to my committee members: Dr. -

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -
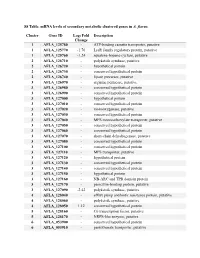
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Structural Properties of the Nickel Ions in Urease: Novel Insights Into the Catalytic and Inhibition Mechanisms
Coordination Chemistry Reviews 190–192 (1999) 331–355 www.elsevier.com/locate/ccr Structural properties of the nickel ions in urease: novel insights into the catalytic and inhibition mechanisms Stefano Ciurli a,*, Stefano Benini b, Wojciech R. Rypniewski b, Keith S. Wilson c, Silvia Miletti a, Stefano Mangani d a Institute of Agricultural Chemistry, Uni6ersity of Bologna, Viale Berti Pichat 10, I-40127 Bologna, Italy b European Molecular Biology Laboratory, c/o DESY, Notkestraße 85, D-22603 Hamburg, Germany c Department of Chemistry, Uni6ersity of York, Heslington, York YO15DD, UK d Department of Chemistry, Uni6ersity of Siena, Pian dei Mantellini 44, I-53100 Siena, Italy Accepted 13 March 1999 Contents Abstract.................................................... 331 1. Biological background ......................................... 332 2. Spectroscopic investigations of the urease active site structure .................. 333 3. Crystallographic studies of the native enzyme ............................ 334 4. Crystallographic studies of urease mutants.............................. 341 5. Crystallographic studies of urease–inhibitor complexes ...................... 345 6. Crystallographic study of a transition state analogue bound to urease.............. 348 7. A novel proposal for the urease mechanism ............................. 350 References .................................................. 353 Abstract This work provides a comprehensive critical summary of urease spectroscopy, crystallogra- phy, inhibitor binding, and site-directed -

Abstracts from the 10Th World Congress for Microcirculation
DOI:10.1111/micc.12246 Abstracts Abstracts from the 10th World Congress for Microcirculation September 25th – 27th, 2015 Kyoto, Japan PLENARY LECTURES vascular neutrophils that constantly patrol the lung and can detect and phagocytose bacteria attached to endothelium suggesting some communication inter-cellular communica- PL1 tion. Clearly much anti-microbial activity occurs in the Imaging the microcirculation in infections vasculature prior to dissemination of bacteria into tissues. P Kubes University of Calgary, Calgary, Canada PL2 Using spinning disk microscopy has allowed us to track pathogens and immune cells in the microcirculation. Mapping oxygen in the brain of awake resting Observing the liver microvasculature revealed numerous mice immune speed bumps that slowed, delayed and even S Charpak prevented bacteria from disseminating to other organs. For Laboratory of Neurophysiology and New Microscopies, Inserm U1128, example, when Borrelia burgdorferi enters the vasculature Paris Descartes University, Paris, France they are immediately caught by the liver vascular macro- The brain is extremely sensitive to hypoxia. Yet, the phage, the Kupffer cells and are engulfed and antigens are physiological values of oxygen concentration in the brain presented on CD1d to resident vascular immune cells remain elusive because high resolution measurements have including invariant Natural Killer T cells (iNKT cells). These only been performed during anesthesia, which affects two iNKT cells receive messages and rapidly make gamma main parameters modulating tissue oxygenation, i.e. interferon to help with immunity. Absence of these events neuronal activity and cerebral blood flow. Using the recent leads to massive dissemination of borrelia especially to joints. finding that measurements of capillary erythrocyte-associ- In the joints the iNKT cells live closely apposed but outside ated transients i.e. -

(12) United States Patent (10) Patent No.: US 9.422,609 B2 Teichberg (45) Date of Patent: Aug
USOO9422609B2 (12) United States Patent (10) Patent No.: US 9.422,609 B2 Teichberg (45) Date of Patent: Aug. 23, 2016 (54) METHODS, COMPOSITIONS AND DEVICES (58) Field of Classification Search FOR MANTAINING CHEMICAL BALANCE CPC ........................ C02F 1/725; C12Y 305/01005 OF CHLORINATED WATER USPC ........................... 210/754; 435/195, 227 231 See application file for complete search history. (75) Inventor: Vivian I. Teichberg, Savyon (IL) (56) References Cited (73) Assignees: Mia Levite, Savyon (IL); Yaar Teichberg, Savyon (IL); Nof Lyle U.S. PATENT DOCUMENTS Teichberg, Savyon (IL) 4,793,935 A * 12/1988 Stillman ............... CO2F 1.5236 21Of727 (*) Notice: Subject to any disclaimer, the term of this 6,673,582 B2 * 1/2004 McTavish ..................... 435/122 patent is extended or adjusted under 35 U.S.C. 154(b) by 1044 days. (Continued) (21) Appl. No.: 12/225.18O FOREIGN PATENT DOCUMENTS y x- - - 9 AU 41971 5, 1979 (22) PCT Filed: Mar. 14, 2007 GB 2025919 1, 1980 (86). PCT No.: PCT/L2007/OOO336 (Continued) S 371 (c)(1) OTHER PUBLICATIONS (2), (4) Date: Sep. 16, 2008 Examiner's Report Dated Oct. 6, 2010 From the Australian Govern (87) PCT Pub. No.: WO2007/107.981 ment, IP Australia Re. Application No. 2007228391. (Continued) PCT Pub. Date: Sep. 27, 2007 (65) Prior Publication Data Primary Examiner — Peter Keyworth (74) Attorney, Agent, or Firm — Browdy and Neimark, US 201OfO270228A1 Oct. 28, 2010 PLLC Related U.S. Application Data (57) ABSTRACT (60) Provisional application No. 60/783,028, filed on Mar. A composition-of-matter for use in water treatment, com 17, 2006. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Early Growth Response 1 Regulates Hematopoietic Support and Proliferation in Human Primary Bone Marrow Stromal Cells
Hematopoiesis SUPPLEMENTARY APPENDIX Early growth response 1 regulates hematopoietic support and proliferation in human primary bone marrow stromal cells Hongzhe Li, 1,2 Hooi-Ching Lim, 1,2 Dimitra Zacharaki, 1,2 Xiaojie Xian, 2,3 Keane J.G. Kenswil, 4 Sandro Bräunig, 1,2 Marc H.G.P. Raaijmakers, 4 Niels-Bjarne Woods, 2,3 Jenny Hansson, 1,2 and Stefan Scheding 1,2,5 1Division of Molecular Hematology, Department of Laboratory Medicine, Lund University, Lund, Sweden; 2Lund Stem Cell Center, Depart - ment of Laboratory Medicine, Lund University, Lund, Sweden; 3Division of Molecular Medicine and Gene Therapy, Department of Labora - tory Medicine, Lund University, Lund, Sweden; 4Department of Hematology, Erasmus MC Cancer Institute, Rotterdam, the Netherlands and 5Department of Hematology, Skåne University Hospital Lund, Skåne, Sweden ©2020 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2019.216648 Received: January 14, 2019. Accepted: July 19, 2019. Pre-published: August 1, 2019. Correspondence: STEFAN SCHEDING - [email protected] Li et al.: Supplemental data 1. Supplemental Materials and Methods BM-MNC isolation Bone marrow mononuclear cells (BM-MNC) from BM aspiration samples were isolated by density gradient centrifugation (LSM 1077 Lymphocyte, PAA, Pasching, Austria) either with or without prior incubation with RosetteSep Human Mesenchymal Stem Cell Enrichment Cocktail (STEMCELL Technologies, Vancouver, Canada) for lineage depletion (CD3, CD14, CD19, CD38, CD66b, glycophorin A). BM-MNCs from fetal long bones and adult hip bones were isolated as reported previously 1 by gently crushing bones (femora, tibiae, fibulae, humeri, radii and ulna) in PBS+0.5% FCS subsequent passing of the cell suspension through a 40-µm filter. -

(12) Patent Application Publication (10) Pub. No.: US 2008/0148432 A1 Abad (43) Pub
US 2008O148432A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2008/0148432 A1 Abad (43) Pub. Date: Jun. 19, 2008 (54) TRANSGENIC PLANTS WITH ENHANCED Publication Classification AGRONOMIC TRAITS (51) Int. Cl. AOIH 5/00 (2006.01) CI2N 5/82 (2006.01) (76) Inventor: Mark Scott Abad, Webster Grove, CI2N 5/04 (2006.01) MO (US) (52) U.S. Cl. ......... 800/279: 800/281; 435/419,435/468; 8OO/320.1 Correspondence Address: (57)57 ABSTRACT MONSANTO COMPANY This invention provides transgenic plant cells with recombi 800 N. LINDBERGHBLVD, ATTENTION: GAIL nant DNA for expression of proteins that are useful for P. WUELLNER, IP PARALEGAL (E2NA) imparting enhanced agronomic trait(s) to transgenic crop ST. LOUIS MO 631.67 9 plants. This invention also provides transgenic plants and 9 progeny seed comprising the transgenic plant cells where the plants are selected for having an enhanced trait selected from the group of traits consisting of enhanced water use effi (21) Appl. No.: 11/374,300 ciency, enhanced cold tolerance, increased yield, enhanced nitrogen use efficiency, enhanced seed protein and enhanced seed oil. Also disclosed are methods for manufacturing trans (22) Filed: Dec. 21, 2005 genic seed and plants with enhanced traits. Patent Application Publication Jun. 19, 2008 Sheet 1 of 3 US 2008/0148432 A1 Figure 1. 41905 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - 127 O2 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -