Comparative Genomics of Bradyrhizobium Japonicum CPAC
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -
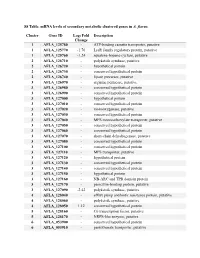
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Genetic and Symbiotic Characterization of Nitrogen-Fixing Bacteria from Three Forest Legumes
BRUNA DANIELA ORTIZ LOPEZ GENETIC AND SYMBIOTIC CHARACTERIZATION OF NITROGEN-FIXING BACTERIA FROM THREE FOREST LEGUMES LAVRAS – MG 2018 BRUNA DANIELA ORTIZ LOPEZ GENETIC AND SYMBIOTIC CHARACTERIZATION OF NITROGEN-FIXING BACTERIA FROM THREE FOREST LEGUMES Dissertação apresentada à Universidade Federal de Lavras, como parte das exigências do Programa de Pós-graduação em Ciência do Solo, área de concentração Biologia, Microbiologia e Processos Biológicos do Solo, para obtenção do título de Mestre. Profa. Dra. Fatima Maria de Souza Moreira Orientadora Dra. Amanda Azarias Guimarães Coorientadora LAVRAS – MG 2018 Ficha catalográfica elaborada pelo Sistema de Geração de Ficha Catalográfica da Biblioteca Universitária da UFLA, com dados informados pelo(a) próprio(a) autor(a). Lopez, Bruna Daniela Ortiz. Genetic and symbiotic characterization of nitrogen-fixing bacteria from three forest legumes / Bruna Daniela Ortiz Lopez. - 2018. 42 p. : il. Orientador(a): Fatima Maria de Souza Moreira. Coorientador(a): Amanda Azarias Guimarães. Dissertação (mestrado acadêmico) - Universidade Federal de Lavras, 2018. Bibliografia. 1. Fixação biológica de nitrogênio. 2. Leguminosas florestais. 3. Caracterização e identificação molecular. I. Moreira, Fatima Maria de Souza. II. Guimarães, Amanda Azarias. III. Título. O conteúdo desta obra é de responsabilidade do(a) autor(a) e de seu orientador(a). BRUNA DANIELA ORTIZ LOPEZ CARACTERIZAÇÃO GENETICA E SIMBIÓTICA DE BACTÉRIAS FIXADORAS DE NITROGÊNIO EM TRÊS LEGUMINOSAS FLORESTAIS GENETIC AND SYMBIOTIC CHARACTERIZATION OF NITROGEN-FIXING BACTERIA FROM THREE FOREST LEGUMES Dissertação apresentada à Universidade Federal de Lavras, como parte das exigências do Programa de Pós-graduação em Ciência do Solo, área de concentração Biologia, Microbiologia e Processos Biológicos do Solo, para obtenção do título de Mestre. -

Phylogeny and Phylogeography of Rhizobial Symbionts Nodulating Legumes of the Tribe Genisteae
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Lincoln University Research Archive G C A T T A C G G C A T genes Review Phylogeny and Phylogeography of Rhizobial Symbionts Nodulating Legumes of the Tribe Genisteae Tomasz St˛epkowski 1,*, Joanna Banasiewicz 1, Camille E. Granada 2, Mitchell Andrews 3 and Luciane M. P. Passaglia 4 1 Autonomous Department of Microbial Biology, Faculty of Agriculture and Biology, Warsaw University of Life Sciences (SGGW), Nowoursynowska 159, 02-776 Warsaw, Poland; [email protected] 2 Universidade do Vale do Taquari—UNIVATES, Rua Avelino Tallini, 171, 95900-000 Lajeado, RS, Brazil; [email protected] 3 Faculty of Agriculture and Life Sciences, Lincoln University, P.O. Box 84, Lincoln 7647, New Zealand; [email protected] 4 Departamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul. Av. Bento Gonçalves, 9500, Caixa Postal 15.053, 91501-970 Porto Alegre, RS, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +48-509-453-708 Received: 31 January 2018; Accepted: 5 March 2018; Published: 14 March 2018 Abstract: The legume tribe Genisteae comprises 618, predominantly temperate species, showing an amphi-Atlantic distribution that was caused by several long-distance dispersal events. Seven out of the 16 authenticated rhizobial genera can nodulate particular Genisteae species. Bradyrhizobium predominates among rhizobia nodulating Genisteae legumes. Bradyrhizobium strains that infect Genisteae species belong to both the Bradyrhizobium japonicum and Bradyrhizobium elkanii superclades. In symbiotic gene phylogenies, Genisteae bradyrhizobia are scattered among several distinct clades, comprising strains that originate from phylogenetically distant legumes. -

Purification and Characterization of a Novel Nitrilase of Rhodococcus
JOURNAL OF BACTERIOLOGY, Sept. 1990, p. 4807-4815 Vol. 172, No. 9 0021-9193/90/094807-09$02.00/0 Copyright X3 1990, American Society for Microbiology Purification and Characterization of a Novel Nitrilase of Rhodococcus rhodochrous K22 That Acts on Aliphatic Nitriles MICHIHIKO KOBAYASHI,* NORIYUKI YANAKA, TORU NAGASAWA, AND HIDEAKI YAMADA Department ofAgricultural Chemistry, Faculty ofAgriculture, Kyoto University, Sakyo-ku, Kyoto 606, Japan Received 18 April 1990/Accepted 8 June 1990 A novel nitrilase that preferentially catalyzes the hydrolysis of aliphatic nitriles to the corresponding carboxylic acids and ammonia was found in the cells of a facultative crotononitrile-utilizing actinomycete isolated from soil. The strain was taxonomically studied and identified as Rhodococcus rhodochrous. The nitrilase was purified, with 9.08% overall recovery, through five steps from a cell extract of the stain. After the last step, the purified enzyme appeared to be homogeneous, as judged by polyacrylamide gel electrophoresis, analytical centrifugation, and double immunodiffusion in agarose. The relative molecular weight values for the native enzyme, estimated from the ultracentrifugal equilibrium and by high-performance liquid chromatog- raphy, were approximately 604,000 + 30,000 and 650,000, respectively, and the enzyme consisted of 15 to 16 subunits identical in molecular weight (41,000). The enzyme acted on aliphatic olefinic nitriles such as crotononitrile and acrylonitrile as the most suitable substrates. The apparent Km values for crotononitrile and acrylonitrile were 18.9 and 1.14 mM, respectively. The nitrilase also catalyzed the direct hydrolysis of saturated aliphatic nitriles, such as valeronitrile, 4-chlorobutyronitrile, and glutaronitrile, to the correspond- ing acids without the formation of amide intermediates. -

Direct Link to Fulltext
ISOLATION, IDENTIFICATION AND SUBSTRATE SPECIFICITY OF A NITRILASE PRODUCING BACTERIA, Acidovorax sp. SK1 Soumya Koippully, Subha Swaraj Pattnaik, Siddhardha Busi* Address(es): Department of Microbiology, School of Life Sciences, Pondicherry University, Puducherry-605 014, India. *Corresponding author: [email protected] doi: 10.15414/jmbfs.2018.8.2.788-793 ARTICLE INFO ABSTRACT Received 13. 5. 2018 The process of biocatalysis or biotransformation remains the core of industrial biotechnology owing to its importance in the synthesis of Revised 3. 9. 2018 high-value products in a cost effective manner. Nitrile biotransformation has also achieved considerable attention in last few decades Accepted 5. 9. 2018 due to its widespread biotechnological and industrial applications. In the present study, a versatile, potent nitrile-degrading bacterium, Published 1. 10. 2018 Acidovorax sp. SK1 was isolated from the cracker waste dumping site of Sivakasi, Tamilnadu and characterized for its biocatalytic potential, nitrilase production and enzyme activity. A pH sensitive indicator-based assay was performed to identify the nitrile degrading ability of the isolated samples. Semi quantitative High performance thin layer chromatographic (HPTLC) method was performed for Regular article quantitative measurement of mandelic acid produced from degradation of nitrile compound, mandelonitrile. The optimization of medium and nutritional parameters were studied for the improvement of nitrilase activity, which indicated that maximum nitrilase activity was observed at an optimum pH of 7.0, agitation at 100 rpm, glucose as best carbon source (10 g/L) and yeast extract (0.1 g/L) as principal nitrogen source. Biomass is also a critical parameter in the biocatalysis of mandelonitrile to mandelic acid and at a biomass of 100 mg/L, maximum nitrilase activity of 0.026 I.U was observed. -

Supplementary Material Supplementary Results And
R. D. Barabote SUPPLEMENTARY MATERIAL SUPPLEMENTARY RESULTS AND DISCUSSION tRNA and codon usage. Forty-five tRNAs representing 43 different anticodons are encoded in the genome (Supplementary Table S1). The tRNAMet is present in three copies in the genome. In contrast to the number of tRNAs, all 61 sense codons are encoded in the genome sequence. The codon usage correlates well with the tRNA complement and is consistent with the high G+C content of the genome as the GC-rich codons predominate in the organism (Supplementary Table S1). Codons ATA (Ile), CGC (Arg), and CGA (Arg) as well as all codons that have a T at the third position, with the exception of CGT (Arg), do not appear to have a cognate tRNA in A. cellulolyticus. As a likely evolutionary adaptation to the available tRNAs for any given amino acid, codons that do not have a cognate tRNA occur with the least frequency in the A. cellulolyticus genome when compared to synonymous codons differing just in the 3rd position. However, the exceptions to these are for glycine (GGT (18.8%) > GGA (14.4%)), leucine (CTT (10.6%) > CTA (1.4%)), arginine (CGC (33.3%) > CGT (11.1%)), and valine (GTT (10.4%) > GTA (3.8%)). The relative preference for CGC codon over CGT codon follows the high G+C content of the genome, while the remaining four biases mentioned above may simply reflect evolutionary conservation of codon usage, as a similar trend is seen in Frankia (Supplementary Table S2). The functional significance of this bias remains elusive. The A. cellulolyticus genome encodes a parsimonious complement of 46 tRNAs. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility. -

Bacteria-Inducing Legume Nodules Involved in the Improvement of Plant Growth, Health and Nutrition
Bacteria-Inducing Legume Nodules Involved in the Improvement of Plant 4 Growth, Health and Nutrition Encarna Velázquez, Lorena Carro, José David Flores-Félix, Esther Menéndez, Martha-Helena Ramírez-Bahena, and Alvaro Peix Abstract Bacteria-inducing legume nodules are known as rhizobia and belong to the class Alphaproteobacteria and Betaproteobacteria. They promote the growth and nutrition of their respective legume hosts through atmospheric nitrogen fixation which takes place in the nodules induced in their roots or stems. In addition, rhizobia have other plant growth-promoting mechanisms, mainly solubilization of phosphate and production of indoleacetic acid, ACC deaminase and sidero- phores. Some of these mechanisms have been reported for strains of rhizobia which are also able to promote the growth of several nonlegumes, such as cere- als, oilseeds and vegetables. Less studied are the mechanisms that have the rhi- zobia to promote the plant health; however, these bacteria are able to exert biocontrol of some phytopathogens and to induce the plant resistance. In this chapter, we revised the available data about the ability of the legume nodule- inducing bacteria for improving the plant growth, health and nutrition of both legumes and nonlegumes. These data showed that rhizobia meet all the require- ments of sustainable agriculture to be used as bio-inoculants allowing the total or partial replacement of chemicals used for fertilization or protection of crops. E. Velázquez (*) Departamento de Microbiología y Genética, Universidad de Salamanca, Salamanca, Spain Unidad Asociada Universidad de Salamanca- CSIC ‘Interacción Planta-Microorganismo’, Salamanca, Spain e-mail: [email protected] L. Carro · J. D. Flores-Félix · E. Menéndez · M.-H. -

Genetic Characterisaton of Rhodococcus Rhodochrous ATCC
The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town Genetic characterization of Rhodococcus rhodochrous ATCC BAA-870 with emphasis on nitrile hydrolysing enzymes n ow Joni Frederick A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy in the Departmentty of of MolecularCape and T Cell Biology, Universitysi of Cape Town er UnivSupervisor: Professor B. T. Sewell Co-supervisor: Professor D. Brady February 2013 Keywords Nitrile hydrolysis Biocatalysis Rhodococcus rhodochrous ATCC BAA-870 Genome sequencing Nitrilase Nitrile hydratase n ow ty of Cape T si er Univ ii Keywords Abstract Rhodococcus rhodochrous ATCC BAA-870 (BAA-870) had previously been isolated on selective media for enrichment of nitrile hydrolysing bacteria. The organism was found to have a wide substrate range, with activity against aliphatics, aromatics, and aryl aliphatics, and enantioselectivity towards beta substituted nitriles and beta amino nitriles, compounds that have potential applications in the pharmaceutical industry. This makes R. rhodochrous ATCC BAA-870 potentially a versatile biocatalyst for the synthesis of a broad range of compounds with amide and carboxylic acid groups that can be derived from structurally related nitrile precursors. The selectivity of biocatalysts allows for high product yields and better atom economyn than non- selective chemical methods of performing this reaction, suchow as acid or base hydrolysis. -

Functional Genomics Approaches to Studying Symbioses Between Legumes and Nitrogen-Fixing Rhizobia
Review Functional Genomics Approaches to Studying Symbioses between Legumes and Nitrogen-Fixing Rhizobia Martina Lardi and Gabriella Pessi * ID Department of Plant and Microbial Biology, University of Zurich, CH-8057 Zurich, Switzerland; [email protected] * Correspondence: [email protected]; Tel.: +41-44-635-2904 Received: 9 April 2018; Accepted: 16 May 2018; Published: 18 May 2018 Abstract: Biological nitrogen fixation gives legumes a pronounced growth advantage in nitrogen- deprived soils and is of considerable ecological and economic interest. In exchange for reduced atmospheric nitrogen, typically given to the plant in the form of amides or ureides, the legume provides nitrogen-fixing rhizobia with nutrients and highly specialised root structures called nodules. To elucidate the molecular basis underlying physiological adaptations on a genome-wide scale, functional genomics approaches, such as transcriptomics, proteomics, and metabolomics, have been used. This review presents an overview of the different functional genomics approaches that have been performed on rhizobial symbiosis, with a focus on studies investigating the molecular mechanisms used by the bacterial partner to interact with the legume. While rhizobia belonging to the alpha-proteobacterial group (alpha-rhizobia) have been well studied, few studies to date have investigated this process in beta-proteobacteria (beta-rhizobia). Keywords: alpha-rhizobia; beta-rhizobia; symbiosis; transcriptomics; proteomics; metabolomics; flavonoids; root exudates; root nodule 1. Introduction Nitrogen fixation in agricultural systems is of enormous agricultural importance, as it increases in situ the fixed nitrogen content of soil and can replace expensive and harmful chemical fertilizers [1]. For over a hundred years, all of the described symbiotic relationships between legumes and nitrogen-fixing rhizobia were confined to the alpha-proteobacteria (alpha-rhizobia) group, which includes Bradyrhizobium, Mesorhizobium, Methylobacterium, Rhizobium, and Sinorhizobium [2]. -

Advances in Biochemistry and Biotechnology Bacterial
Advances in Biochemistry and Biotechnology Degrassi G and Carpentieri-Pipolo V. Adv Biochem Biotechnol 5: 1099. Review Article DOI: 10.29011/2574-7258.001099 Bacterial Endophytes Associated to Crops: Novel Practices for Sustainable Agriculture Giuliano Degrassi1*, Valeria Carpentieri-Pipolo2 1Industrial Biotechnology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), Parque Tecnológico Miguelete, San Martín, Buenos Aires, República Argentina 2Embrapa Trigo, Rodovia BR-285, Km 294, 99050-970, Passo Fundo, Rio Grande do Sul, Brazil. *Corresponding author: Giuliano Degrassi, Industrial Biotechnology Group, International Centre for Genetic Engineering and Bio- technology (ICGEB), Parque Tecnológico Miguelete, Av. General Paz N° 5445, B1650WAB San Martín, Buenos Aires, República Argentina Citation: Degrassi G and Carpentieri-Pipolo V (2020) Bacterial Endophytes Associated to Crops: Novel Practices for Sustainable Agriculture. Adv Biochem Biotechnol 5: 1099. DOI: 10.29011/2574-7258.001099 Received Date: 16 April, 2020; Accepted Date: 20 May, 2020; Published Date: 25 May, 2020 Abstract For many decades, rhizosphere bacteria have been studied for their potential to promote crop growth and control certain pathogens. Compared to the studies conducted, there are relatively few examples of microbial inoculants based on rhizosphere bacteria that have had commercial success, mainly due to the strong competition present in the rhizosphere. In the last twenty years, studies on endophytes have multiplied, also and above all as a possible alternative to rhizobacteria, for the development of microbial inoculants capable of replacing some agrochemicals and reducing the environmental impact of agronomic management of crops. This minireview summarizes the most important characteristics and qualities of endophytic bacteria and describes the path that can be followed to identify and deepen the knowledge of candidates suitable for the development of microbial inocu- lants.