Genetic Characterisaton of Rhodococcus Rhodochrous ATCC
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -
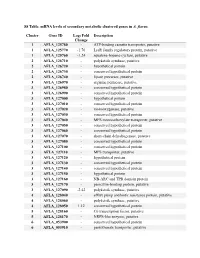
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Comparative Genomics of Bradyrhizobium Japonicum CPAC
Siqueira et al. BMC Genomics 2014, 15:420 http://www.biomedcentral.com/1471-2164/15/420 RESEARCH ARTICLE Open Access Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean Arthur Fernandes Siqueira1,2†, Ernesto Ormeño-Orrillo3†,RangelCelsoSouza4,ElisetePainsRodrigues5, Luiz Gonzaga Paula Almeida4, Fernando Gomes Barcellos5, Jesiane Stefânia Silva Batista6, Andre Shigueyoshi Nakatani2, Esperanza Martínez-Romero3, Ana Tereza Ribeiro Vasconcelos4 and Mariangela Hungria1,2* Abstract Background: The soybean-Bradyrhizobium symbiosis can be highly efficient in fixing nitrogen, but few genomic sequences of elite inoculant strains are available. Here we contribute with information on the genomes of two commercial strains that are broadly applied to soybean crops in the tropics. B. japonicum CPAC 15 (=SEMIA 5079) is outstanding in its saprophytic capacity and competitiveness, whereas B. diazoefficiens CPAC 7 (=SEMIA 5080) is known for its high efficiency in fixing nitrogen. Both are well adapted to tropical soils. The genomes of CPAC 15 and CPAC 7 were compared to each other and also to those of B. japonicum USDA 6T and B. diazoefficiens USDA 110T. Results: Differences in genome size were found between species, with B. japonicum having larger genomes than B. diazoefficiens. Although most of the four genomes were syntenic, genome rearrangements within and between species were observed, including events in the symbiosis island. In addition to the symbiotic region, several genomic islands were identified. Altogether, these features must confer high genomic plasticity that might explain adaptation and differences in symbiotic performance. It was not possible to attribute known functions to half of the predicted genes. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species
pathogens Review Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species Irina B. Ivshina 1,2,*, Maria S. Kuyukina 1,2 , Anastasiia V. Krivoruchko 1,2 and Elena A. Tyumina 1,2 1 Perm Federal Research Center UB RAS, Institute of Ecology and Genetics of Microorganisms UB RAS, 13 Golev Str., 614081 Perm, Russia; [email protected] (M.S.K.); [email protected] (A.V.K.); [email protected] (E.A.T.) 2 Department of Microbiology and Immunology, Perm State University, 15 Bukirev Str., 614990 Perm, Russia * Correspondence: [email protected]; Tel.: +7-342-280-8114 Abstract: Under conditions of increasing environmental pollution, true saprophytes are capable of changing their survival strategies and demonstrating certain pathogenicity factors. Actinobacteria of the genus Rhodococcus, typical soil and aquatic biotope inhabitants, are characterized by high ecological plasticity and a wide range of oxidized organic substrates, including hydrocarbons and their derivatives. Their cell adaptations, such as the ability of adhering and colonizing surfaces, a complex life cycle, formation of resting cells and capsule-like structures, diauxotrophy, and a rigid cell wall, developed against the negative effects of anthropogenic pollutants are discussed and the risks of possible pathogenization of free-living saprotrophic Rhodococcus species are proposed. Due to universal adaptation features, Rhodococcus species are among the candidates, if further anthropogenic pressure increases, to move into the group of potentially pathogenic organisms with “unprofessional” parasitism, and to join an expanding list of infectious agents as facultative or occasional parasites. Citation: Ivshina, I.B.; Kuyukina, Keywords: actinobacteria; Rhodococcus; pathogenicity factors; adhesion; autoaggregation; colonization; M.S.; Krivoruchko, A.V.; Tyumina, defense against phagocytosis; adaptive strategies E.A. -

Purification and Characterization of a Novel Nitrilase of Rhodococcus
JOURNAL OF BACTERIOLOGY, Sept. 1990, p. 4807-4815 Vol. 172, No. 9 0021-9193/90/094807-09$02.00/0 Copyright X3 1990, American Society for Microbiology Purification and Characterization of a Novel Nitrilase of Rhodococcus rhodochrous K22 That Acts on Aliphatic Nitriles MICHIHIKO KOBAYASHI,* NORIYUKI YANAKA, TORU NAGASAWA, AND HIDEAKI YAMADA Department ofAgricultural Chemistry, Faculty ofAgriculture, Kyoto University, Sakyo-ku, Kyoto 606, Japan Received 18 April 1990/Accepted 8 June 1990 A novel nitrilase that preferentially catalyzes the hydrolysis of aliphatic nitriles to the corresponding carboxylic acids and ammonia was found in the cells of a facultative crotononitrile-utilizing actinomycete isolated from soil. The strain was taxonomically studied and identified as Rhodococcus rhodochrous. The nitrilase was purified, with 9.08% overall recovery, through five steps from a cell extract of the stain. After the last step, the purified enzyme appeared to be homogeneous, as judged by polyacrylamide gel electrophoresis, analytical centrifugation, and double immunodiffusion in agarose. The relative molecular weight values for the native enzyme, estimated from the ultracentrifugal equilibrium and by high-performance liquid chromatog- raphy, were approximately 604,000 + 30,000 and 650,000, respectively, and the enzyme consisted of 15 to 16 subunits identical in molecular weight (41,000). The enzyme acted on aliphatic olefinic nitriles such as crotononitrile and acrylonitrile as the most suitable substrates. The apparent Km values for crotononitrile and acrylonitrile were 18.9 and 1.14 mM, respectively. The nitrilase also catalyzed the direct hydrolysis of saturated aliphatic nitriles, such as valeronitrile, 4-chlorobutyronitrile, and glutaronitrile, to the correspond- ing acids without the formation of amide intermediates. -

Direct Link to Fulltext
ISOLATION, IDENTIFICATION AND SUBSTRATE SPECIFICITY OF A NITRILASE PRODUCING BACTERIA, Acidovorax sp. SK1 Soumya Koippully, Subha Swaraj Pattnaik, Siddhardha Busi* Address(es): Department of Microbiology, School of Life Sciences, Pondicherry University, Puducherry-605 014, India. *Corresponding author: [email protected] doi: 10.15414/jmbfs.2018.8.2.788-793 ARTICLE INFO ABSTRACT Received 13. 5. 2018 The process of biocatalysis or biotransformation remains the core of industrial biotechnology owing to its importance in the synthesis of Revised 3. 9. 2018 high-value products in a cost effective manner. Nitrile biotransformation has also achieved considerable attention in last few decades Accepted 5. 9. 2018 due to its widespread biotechnological and industrial applications. In the present study, a versatile, potent nitrile-degrading bacterium, Published 1. 10. 2018 Acidovorax sp. SK1 was isolated from the cracker waste dumping site of Sivakasi, Tamilnadu and characterized for its biocatalytic potential, nitrilase production and enzyme activity. A pH sensitive indicator-based assay was performed to identify the nitrile degrading ability of the isolated samples. Semi quantitative High performance thin layer chromatographic (HPTLC) method was performed for Regular article quantitative measurement of mandelic acid produced from degradation of nitrile compound, mandelonitrile. The optimization of medium and nutritional parameters were studied for the improvement of nitrilase activity, which indicated that maximum nitrilase activity was observed at an optimum pH of 7.0, agitation at 100 rpm, glucose as best carbon source (10 g/L) and yeast extract (0.1 g/L) as principal nitrogen source. Biomass is also a critical parameter in the biocatalysis of mandelonitrile to mandelic acid and at a biomass of 100 mg/L, maximum nitrilase activity of 0.026 I.U was observed. -

Structures, Functions, and Mechanisms of Filament Forming Enzymes: a Renaissance of Enzyme Filamentation
Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation A Review By Chad K. Park & Nancy C. Horton Department of Molecular and Cellular Biology University of Arizona Tucson, AZ 85721 N. C. Horton ([email protected], ORCID: 0000-0003-2710-8284) C. K. Park ([email protected], ORCID: 0000-0003-1089-9091) Keywords: Enzyme, Regulation, DNA binding, Nuclease, Run-On Oligomerization, self-association 1 Abstract Filament formation by non-cytoskeletal enzymes has been known for decades, yet only relatively recently has its wide-spread role in enzyme regulation and biology come to be appreciated. This comprehensive review summarizes what is known for each enzyme confirmed to form filamentous structures in vitro, and for the many that are known only to form large self-assemblies within cells. For some enzymes, studies describing both the in vitro filamentous structures and cellular self-assembly formation are also known and described. Special attention is paid to the detailed structures of each type of enzyme filament, as well as the roles the structures play in enzyme regulation and in biology. Where it is known or hypothesized, the advantages conferred by enzyme filamentation are reviewed. Finally, the similarities, differences, and comparison to the SgrAI system are also highlighted. 2 Contents INTRODUCTION…………………………………………………………..4 STRUCTURALLY CHARACTERIZED ENZYME FILAMENTS…….5 Acetyl CoA Carboxylase (ACC)……………………………………………………………………5 Phosphofructokinase (PFK)……………………………………………………………………….6 -

Production of Muconic Acid in Plants T ⁎ Aymerick Eudesa,B, , Roland Berthomieua,C, Zhangying Haoa,B, Nanxia Zhaoa,D, ⁎ Veronica Teixeira Benitesa,E, Edward E.K
Metabolic Engineering 46 (2018) 13–19 Contents lists available at ScienceDirect Metabolic Engineering journal homepage: www.elsevier.com/locate/meteng Production of muconic acid in plants T ⁎ Aymerick Eudesa,b, , Roland Berthomieua,c, Zhangying Haoa,b, Nanxia Zhaoa,d, ⁎ Veronica Teixeira Benitesa,e, Edward E.K. Baidooa,e, Dominique Loquéa,b,f,g, a Joint BioEnergy Institute, EmeryStation East, 5885 Hollis St, 4th Floor, Emeryville, CA 94608, USA b Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA c Ecole Polytechnique, Université Paris-Saclay, Palaiseau 91120, France d Department of Bioengineering, Department of Chemical & Biomolecular Engineering, University of California, Berkeley, CA 94720, USA e Biological Systems and Engineering Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA f Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720, USA g Université Lyon 1, INSA de Lyon, CNRS, UMR5240, Microbiologie, Adaptation et Pathogénie, 10 rue Raphaël Dubois, F-69622, Villeurbanne, France ARTICLE INFO ABSTRACT Keywords: Muconic acid (MA) is a dicarboxylic acid used for the production of industrially relevant chemicals such as Muconic acid adipic acid, terephthalic acid, and caprolactam. Because the synthesis of these polymer precursors generates Salicylic acid toxic intermediates by utilizing petroleum-derived chemicals and corrosive catalysts, the development of al- Catechol ternative strategies for the bio-based production of MA has garnered significant interest. Plants produce organic Shikimate carbon skeletons by harvesting carbon dioxide and energy from the sun, and therefore represent advantageous Plastid hosts for engineered metabolic pathways towards the manufacturing of chemicals. -

Bacterial Degradation of Isoprene in the Terrestrial Environment Myriam
Bacterial Degradation of Isoprene in the Terrestrial Environment Myriam El Khawand A thesis submitted to the University of East Anglia in fulfillment of the requirements for the degree of Doctor of Philosophy School of Environmental Sciences November 2014 ©This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derive there from must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. Abstract Isoprene is a climate active gas emitted from natural and anthropogenic sources in quantities equivalent to the global methane flux to the atmosphere. 90 % of the emitted isoprene is produced enzymatically in the chloroplast of terrestrial plants from dimethylallyl pyrophosphate via the methylerythritol pathway. The main role of isoprene emission by plants is to reduce the damage caused by heat stress through stabilizing cellular membranes. Isoprene emission from microbes, animals, and humans has also been reported, albeit less understood than isoprene emission from plants. Despite large emissions, isoprene is present at low concentrations in the atmosphere due to its rapid reactions with other atmospheric components, such as hydroxyl radicals. Isoprene can extend the lifetime of potent greenhouse gases, influence the tropospheric concentrations of ozone, and induce the formation of secondary organic aerosols. While substantial knowledge exists about isoprene production and atmospheric chemistry, our knowledge of isoprene sinks is limited. Soils consume isoprene at a high rate and contain numerous isoprene-utilizing bacteria. However, Rhodococcus sp. AD45 is the only terrestrial isoprene-degrading bacterium characterized in any detail. -

429 Chem. Listy 97, 363–520 (2003)
Chem. Listy 97, 363–520 (2003) Posters P117 APPLICATION OF DEGENERATE REFERENCES OLIGONUCLEOTIDE GENE SHUFFLING FOR CONSTRUCTION OF HYBRID 1. Janssen D. B., Pries F., Ploeg J., Kazemier B., Terpstra P., HALOALKANE DEHALOGENASE Witholt B.: J. Bacteriol. 171, 6791 (1989). 2. Nagata Y., Nariya T., Ohtomo R., Fukuda M., Yano K., ANDREA JESENSKÁa, YUJI NAGATAb, Takagi M.: J. Bacteriol. 175, 6403 (1993). and JIŘÍ DAMBORSKÝc 3. Kulakova A. N., Larkin M. J., Kulakov L. A.: Microbio- logy 143, 109 (1997). a National Centre for Biomolecular Research, Faculty of Science, 4. Jesenska A., Bartos M., Czernekova V., Rychlik I., Masaryk University, Kotlářská 2, Brno, 611 37, Czech Republic, Pavlik I., Damborsky V.: Appl. Environ. Microbiol. 68, e-mail: [email protected]; bDepartment of Environmen- 3724 (2002). tal Life Sciences, Graduate School of Life Sciences, Tohoku 5. Gibbs M. D., Nevalainen K. M., Bergquist P. L.: Gene University, Sendai 980-8577, Japan 271, 13 (2001). Keywords: haloalkane dehalogenase, in vitro recombination, P118 KINETICS AND SPECIFICITY construct, substrate specificity OF HALOALKANE DEHALOGENASE LinB FROM Sphingomonas paucimobilis UT26 Halogenated organic compounds constitute one of the ZBYNĚK PROKOPa, MARTA MONINCOVÁa, largest groups of environmental pollutants as a result of their MARTIN KLVAŇAa, RADKA CHALOUPKOVÁa, widespread use in industry and agriculture. Haloalkane de- DICK B. JANSSENb, YUJI NAGATAc, halogenases hydrolytically convert halogenated aliphatic and JIŘÍ DAMBORSKÝa compounds to corresponding alcohols. Enhancement of cata- lytic properties of environmentally important enzymes using aNational Centre for Biomolecular Research, Masaryk Univer- in vitro evolution techniques is one of the obvious goal of bio- sity, Kotlářská 2, 611 37 Brno, Czech Republic; bUniversity of technology.