Purification and Characterization of a Novel Nitrilase of Rhodococcus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mechanistic Insights Into the Hydrocyanation Reaction
Mechanistic insights into the hydrocyanation reaction Citation for published version (APA): Bini, L. (2009). Mechanistic insights into the hydrocyanation reaction. Technische Universiteit Eindhoven. https://doi.org/10.6100/IR644067 DOI: 10.6100/IR644067 Document status and date: Published: 01/01/2009 Document Version: Publisher’s PDF, also known as Version of Record (includes final page, issue and volume numbers) Please check the document version of this publication: • A submitted manuscript is the version of the article upon submission and before peer-review. There can be important differences between the submitted version and the official published version of record. People interested in the research are advised to contact the author for the final version of the publication, or visit the DOI to the publisher's website. • The final author version and the galley proof are versions of the publication after peer review. • The final published version features the final layout of the paper including the volume, issue and page numbers. Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal. -

Numerical Simulation of a Two-Phase Flow for the Acrylonitrile Electrolytic Adiponitrile Process in a Vertical/Horizontal Electrolysis Cell
energies Article Numerical Simulation of a Two-Phase Flow for the Acrylonitrile Electrolytic Adiponitrile Process in a Vertical/Horizontal Electrolysis Cell Jiin-Yuh Jang * and Yu-Feng Gan Department of Mechanical Engineering, National Cheng-Kung University, Tainan 70101, Taiwan; [email protected] * Correspondence: [email protected]; Tel.: +886-6-2088573 Received: 7 September 2018; Accepted: 6 October 2018; Published: 12 October 2018 Abstract: This paper investigated the effect of oxygen holdup on the current density distribution over the electrode of a vertical/horizontal electrolysis cell with a two-dimensional Eulerian–Eulerian two-phase flow model in the acrylonitrile (AN) electrolytic adiponitrile (ADN) process. The physical models consisted of a vertical/horizontal electrolysis cell 10 mm wide and 600 mm long. The electrical potential difference between the anode and cathode was fixed at 5 V, which corresponded to a uniform current density j = 0.4 A/cm2 without any bubbles released from the electrodes. The effects of different inlet electrolyte velocities (vin = 0.4, 0.6, 1.0 and 1.5 m/s) on the void fraction and the current density distributions were discussed in detail. It is shown that, for a given applied voltage, as the electrolyte velocity is increased, the gas diffusion layer thickness decreased and this resulted in the decrease of the gas void fraction and increase of the corresponding current density; for a given velocity, the current density for a vertical cell was higher than that for a horizontal cell. Furthermore, assuming the release of uniform mass flux for the oxygen results in overestimation of the total gas accumulation mass flow rate by 2.8% and 5.8% and it will also result in underestimation of the current density by 0.3% and 2.4% for a vertical cell and a horizontal cell, respectively. -

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

Transport of Dangerous Goods
ST/SG/AC.10/1/Rev.16 (Vol.I) Recommendations on the TRANSPORT OF DANGEROUS GOODS Model Regulations Volume I Sixteenth revised edition UNITED NATIONS New York and Geneva, 2009 NOTE The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the Secretariat of the United Nations concerning the legal status of any country, territory, city or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. ST/SG/AC.10/1/Rev.16 (Vol.I) Copyright © United Nations, 2009 All rights reserved. No part of this publication may, for sales purposes, be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, electrostatic, magnetic tape, mechanical, photocopying or otherwise, without prior permission in writing from the United Nations. UNITED NATIONS Sales No. E.09.VIII.2 ISBN 978-92-1-139136-7 (complete set of two volumes) ISSN 1014-5753 Volumes I and II not to be sold separately FOREWORD The Recommendations on the Transport of Dangerous Goods are addressed to governments and to the international organizations concerned with safety in the transport of dangerous goods. The first version, prepared by the United Nations Economic and Social Council's Committee of Experts on the Transport of Dangerous Goods, was published in 1956 (ST/ECA/43-E/CN.2/170). In response to developments in technology and the changing needs of users, they have been regularly amended and updated at succeeding sessions of the Committee of Experts pursuant to Resolution 645 G (XXIII) of 26 April 1957 of the Economic and Social Council and subsequent resolutions. -
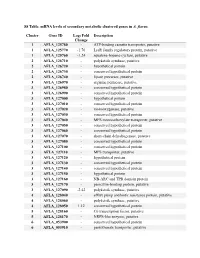
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Comparative Genomics of Bradyrhizobium Japonicum CPAC
Siqueira et al. BMC Genomics 2014, 15:420 http://www.biomedcentral.com/1471-2164/15/420 RESEARCH ARTICLE Open Access Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean Arthur Fernandes Siqueira1,2†, Ernesto Ormeño-Orrillo3†,RangelCelsoSouza4,ElisetePainsRodrigues5, Luiz Gonzaga Paula Almeida4, Fernando Gomes Barcellos5, Jesiane Stefânia Silva Batista6, Andre Shigueyoshi Nakatani2, Esperanza Martínez-Romero3, Ana Tereza Ribeiro Vasconcelos4 and Mariangela Hungria1,2* Abstract Background: The soybean-Bradyrhizobium symbiosis can be highly efficient in fixing nitrogen, but few genomic sequences of elite inoculant strains are available. Here we contribute with information on the genomes of two commercial strains that are broadly applied to soybean crops in the tropics. B. japonicum CPAC 15 (=SEMIA 5079) is outstanding in its saprophytic capacity and competitiveness, whereas B. diazoefficiens CPAC 7 (=SEMIA 5080) is known for its high efficiency in fixing nitrogen. Both are well adapted to tropical soils. The genomes of CPAC 15 and CPAC 7 were compared to each other and also to those of B. japonicum USDA 6T and B. diazoefficiens USDA 110T. Results: Differences in genome size were found between species, with B. japonicum having larger genomes than B. diazoefficiens. Although most of the four genomes were syntenic, genome rearrangements within and between species were observed, including events in the symbiosis island. In addition to the symbiotic region, several genomic islands were identified. Altogether, these features must confer high genomic plasticity that might explain adaptation and differences in symbiotic performance. It was not possible to attribute known functions to half of the predicted genes. -

Direct Link to Fulltext
ISOLATION, IDENTIFICATION AND SUBSTRATE SPECIFICITY OF A NITRILASE PRODUCING BACTERIA, Acidovorax sp. SK1 Soumya Koippully, Subha Swaraj Pattnaik, Siddhardha Busi* Address(es): Department of Microbiology, School of Life Sciences, Pondicherry University, Puducherry-605 014, India. *Corresponding author: [email protected] doi: 10.15414/jmbfs.2018.8.2.788-793 ARTICLE INFO ABSTRACT Received 13. 5. 2018 The process of biocatalysis or biotransformation remains the core of industrial biotechnology owing to its importance in the synthesis of Revised 3. 9. 2018 high-value products in a cost effective manner. Nitrile biotransformation has also achieved considerable attention in last few decades Accepted 5. 9. 2018 due to its widespread biotechnological and industrial applications. In the present study, a versatile, potent nitrile-degrading bacterium, Published 1. 10. 2018 Acidovorax sp. SK1 was isolated from the cracker waste dumping site of Sivakasi, Tamilnadu and characterized for its biocatalytic potential, nitrilase production and enzyme activity. A pH sensitive indicator-based assay was performed to identify the nitrile degrading ability of the isolated samples. Semi quantitative High performance thin layer chromatographic (HPTLC) method was performed for Regular article quantitative measurement of mandelic acid produced from degradation of nitrile compound, mandelonitrile. The optimization of medium and nutritional parameters were studied for the improvement of nitrilase activity, which indicated that maximum nitrilase activity was observed at an optimum pH of 7.0, agitation at 100 rpm, glucose as best carbon source (10 g/L) and yeast extract (0.1 g/L) as principal nitrogen source. Biomass is also a critical parameter in the biocatalysis of mandelonitrile to mandelic acid and at a biomass of 100 mg/L, maximum nitrilase activity of 0.026 I.U was observed. -

Supplementary Material Supplementary Results And
R. D. Barabote SUPPLEMENTARY MATERIAL SUPPLEMENTARY RESULTS AND DISCUSSION tRNA and codon usage. Forty-five tRNAs representing 43 different anticodons are encoded in the genome (Supplementary Table S1). The tRNAMet is present in three copies in the genome. In contrast to the number of tRNAs, all 61 sense codons are encoded in the genome sequence. The codon usage correlates well with the tRNA complement and is consistent with the high G+C content of the genome as the GC-rich codons predominate in the organism (Supplementary Table S1). Codons ATA (Ile), CGC (Arg), and CGA (Arg) as well as all codons that have a T at the third position, with the exception of CGT (Arg), do not appear to have a cognate tRNA in A. cellulolyticus. As a likely evolutionary adaptation to the available tRNAs for any given amino acid, codons that do not have a cognate tRNA occur with the least frequency in the A. cellulolyticus genome when compared to synonymous codons differing just in the 3rd position. However, the exceptions to these are for glycine (GGT (18.8%) > GGA (14.4%)), leucine (CTT (10.6%) > CTA (1.4%)), arginine (CGC (33.3%) > CGT (11.1%)), and valine (GTT (10.4%) > GTA (3.8%)). The relative preference for CGC codon over CGT codon follows the high G+C content of the genome, while the remaining four biases mentioned above may simply reflect evolutionary conservation of codon usage, as a similar trend is seen in Frankia (Supplementary Table S2). The functional significance of this bias remains elusive. The A. cellulolyticus genome encodes a parsimonious complement of 46 tRNAs. -

Chemical Name Federal P Code CAS Registry Number Acutely
Acutely / Extremely Hazardous Waste List Federal P CAS Registry Acutely / Extremely Chemical Name Code Number Hazardous 4,7-Methano-1H-indene, 1,4,5,6,7,8,8-heptachloro-3a,4,7,7a-tetrahydro- P059 76-44-8 Acutely Hazardous 6,9-Methano-2,4,3-benzodioxathiepin, 6,7,8,9,10,10- hexachloro-1,5,5a,6,9,9a-hexahydro-, 3-oxide P050 115-29-7 Acutely Hazardous Methanimidamide, N,N-dimethyl-N'-[2-methyl-4-[[(methylamino)carbonyl]oxy]phenyl]- P197 17702-57-7 Acutely Hazardous 1-(o-Chlorophenyl)thiourea P026 5344-82-1 Acutely Hazardous 1-(o-Chlorophenyl)thiourea 5344-82-1 Extremely Hazardous 1,1,1-Trichloro-2, -bis(p-methoxyphenyl)ethane Extremely Hazardous 1,1a,2,2,3,3a,4,5,5,5a,5b,6-Dodecachlorooctahydro-1,3,4-metheno-1H-cyclobuta (cd) pentalene, Dechlorane Extremely Hazardous 1,1a,3,3a,4,5,5,5a,5b,6-Decachloro--octahydro-1,2,4-metheno-2H-cyclobuta (cd) pentalen-2- one, chlorecone Extremely Hazardous 1,1-Dimethylhydrazine 57-14-7 Extremely Hazardous 1,2,3,4,10,10-Hexachloro-6,7-epoxy-1,4,4,4a,5,6,7,8,8a-octahydro-1,4-endo-endo-5,8- dimethanonaph-thalene Extremely Hazardous 1,2,3-Propanetriol, trinitrate P081 55-63-0 Acutely Hazardous 1,2,3-Propanetriol, trinitrate 55-63-0 Extremely Hazardous 1,2,4,5,6,7,8,8-Octachloro-4,7-methano-3a,4,7,7a-tetra- hydro- indane Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]- 51-43-4 Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]-, P042 51-43-4 Acutely Hazardous 1,2-Dibromo-3-chloropropane 96-12-8 Extremely Hazardous 1,2-Propylenimine P067 75-55-8 Acutely Hazardous 1,2-Propylenimine 75-55-8 Extremely Hazardous 1,3,4,5,6,7,8,8-Octachloro-1,3,3a,4,7,7a-hexahydro-4,7-methanoisobenzofuran Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime 26419-73-8 Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime. -

BUTADIENE AS a CHEMICAL RAW MATERIAL (September 1998)
Abstract Process Economics Program Report 35D BUTADIENE AS A CHEMICAL RAW MATERIAL (September 1998) The dominant technology for producing butadiene (BD) is the cracking of naphtha to pro- duce ethylene. BD is obtained as a coproduct. As the growth of ethylene production outpaced the growth of BD demand, an oversupply of BD has been created. This situation provides the incen- tive for developing technologies with BD as the starting material. The objective of this report is to evaluate the economics of BD-based routes and to compare the economics with those of cur- rently commercial technologies. In addition, this report addresses commercial aspects of the butadiene industry such as supply/demand, BD surplus, price projections, pricing history, and BD value in nonchemical applications. We present process economics for two technologies: • Cyclodimerization of BD leading to ethylbenzene (DSM-Chiyoda) • Hydrocyanation of BD leading to caprolactam (BASF). Furthermore, we present updated economics for technologies evaluated earlier by PEP: • Cyclodimerization of BD leading to styrene (Dow) • Carboalkoxylation of BD leading to caprolactam and to adipic acid • Hydrocyanation of BD leading to hexamethylenediamine. We also present a comparison of the DSM-Chiyoda and Dow technologies for producing sty- rene. The Dow technology produces styrene directly and is limited in terms of capacity by the BD available from a world-scale naphtha cracker. The 250 million lb/yr (113,000 t/yr) capacity se- lected for the Dow technology requires the BD output of two world-scale naphtha crackers. The DSM-Chiyoda technology produces ethylbenzene. In our evaluations, we assumed a scheme whereby ethylbenzene from a 266 million lb/yr (121,000 t/yr) DSM-Chiyoda unit is combined with 798 million lb/yr (362,000 t/yr) of ethylbenzene produced by conventional alkylation of benzene with ethylene. -

Purification and Characterization of Nitrilase from Fusarium Solani
Process Biochemistry 45 (2010) 1115–1120 Contents lists available at ScienceDirect Process Biochemistry journal homepage: www.elsevier.com/locate/procbio Purification and characterization of nitrilase from Fusarium solani IMI196840 Vojtechˇ Vejvoda a, David Kubácˇ a,Alzbˇ etaˇ Davidová a, Ondrejˇ Kaplan a, Miroslav Sulcˇ a,b, Ondrejˇ Svedaˇ a, Radka Chaloupková c, Ludmila Martínková a,∗ a Institute of Microbiology, Centre of Biocatalysis and Biotransformation, Academy of Sciences of the Czech Republic, Vídenskᡠ1083, CZ-142 20 Prague, Czech Republic b Department of Biochemistry, Faculty of Science, Charles University Prague, Hlavova 8, CZ-128 40 Prague, Czech Republic c Loschmidt Laboratories, Department of Experimental Biology and Centre of Biocatalysis and Biotransformation, Faculty of Science, Masaryk University, Kamenice 5/A4, CZ-625 00 Brno, Czech Republic article info abstract Article history: Nitrilase activity in Fusarium solani IMI196840 (approx. 1500 U l−1 of culture broth) was induced by 2- Received 11 November 2009 cyanopyridine. The enzyme was purified by a factor of 20.3 at a yield of 26.9%. According to gel filtration, Received in revised form 26 March 2010 the holoenzyme was an approx. 550-kDa homooligomer consisting of subunits with a molecular weight Accepted 30 March 2010 of approximately 40 kDa, as determined by SDS-PAGE. Mass spectrometry analysis of the tryptic frag- ments suggested a high similarity of this enzyme to the hypothetical CN hydrolases from Aspergillus Keywords: oryzae, Gibberella zeae, Gibberella moniliformis and Nectria haematococca. Circular dichroism and fluo- Fusarium solani rescence spectra indicated that secondary structure content and overall tertiary structure, respectively, Nitrilase were almost identical in nitrilases from F. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility.