Metabolism and Host Specificity in the Rhizobium Leguminosarum Species Complex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

The Genomes of Polyextremophilic Cyanidiales Contain 1% 2 Horizontally Transferred Genes with Diverse Adaptive Functions 3 4 Alessandro W
bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The genomes of polyextremophilic Cyanidiales contain 1% 2 horizontally transferred genes with diverse adaptive functions 3 4 Alessandro W. Rossoni1#, Dana C. Price2, Mark Seger3, Dagmar Lyska1, Peter Lammers3, 5 Debashish Bhattacharya4 & Andreas P.M. Weber1* 6 7 1Institute of Plant Biochemistry, Cluster of Excellence on Plant Sciences (CEPLAS), Heinrich 8 Heine University, Universitätsstraße 1, 40225 Düsseldorf, Germany 9 2Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA 10 3Arizona Center for Algae Technology and Innovation, Arizona State University, Mesa, AZ 11 85212, USA 12 4Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ 13 08901, USA 14 15 *Corresponding author: Prof. Dr. Andreas P.M. Weber, 16 e-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/526111; this version posted January 23, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics 20 that impact our understanding regarding the origin of metabolic processes and the role of 21 organelles in cellular evolution. -

Pfc5813.Pdf (9.887Mb)
UNIVERSIDAD POLITÉCNICA DE CARTAGENA ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA DEPARTAMENTO DE PRODUCCIÓN VEGETAL INGENIERO AGRÓNOMO PROYECTO FIN DE CARRERA: “AISLAMIENTO E IDENTIFICACIÓN DE LOS RIZOBIOS ASOCIADOS A LOS NÓDULOS DE ASTRAGALUS NITIDIFLORUS”. Realizado por: Noelia Real Giménez Dirigido por: María José Vicente Colomer Francisco José Segura Carreras Cartagena, Julio de 2014. ÍNDICE GENERAL 1. Introducción…………………………………………………….…………………………………………………1 1.1. Astragalus nitidiflorus………………………………..…………………………………………………2 1.1.1. Encuadre taxonómico……………………………….…..………………………………………………2 1.1.2. El origen de Astragalus nitidiflorus………………………………………………………………..4 1.1.3. Descripción de la especie………..…………………………………………………………………….5 1.1.4. Biología…………………………………………………………………………………………………………7 1.1.4.1. Ciclo vegetativo………………….……………………………………………………………………7 1.1.4.2. Fenología de la floración……………………………………………………………………….9 1.1.4.3. Sistema de reproducción……………………………………………………………………….10 1.1.4.4. Dispersión de los frutos…………………………………….…………………………………..11 1.1.4.5. Nodulación con Rhizobium…………………………………………………………………….12 1.1.4.6. Diversidad genética……………………………………………………………………………....13 1.1.5. Ecología………………………………………………………………………………………………..…….14 1.1.6. Corología y tamaño poblacional……………………………………………………..…………..15 1.1.7. Protección…………………………………………………………………………………………………..18 1.1.8. Amenazas……………………………………………………………………………………………………19 1.1.8.1. Factores bióticos…………………………………………………………………………………..19 1.1.8.2. Factores abióticos………………………………………………………………………………….20 1.1.8.3. Factores antrópicos………………..…………………………………………………………….21 -

A Survey of Agrobacterium Strains Associated with Georgia Pecan Trees and an Immunological Study of the Bacterium
AN ABSTRACT OF THE THESIS OF Hacene Bouzar for the degree of Master of Science in Botany & Plant Pathology presented on December 17, 1982 Title: A SURVEY OF AGROBACTERIUM STRAINS ASSOCIATED WITH GEORGIA PECAN TREES AND AN IMMUNOLOGICAL STUDY OF THE BACTERIUM Abstract approved: Redacted for Privacy Larry W. Moore Crown gall was found in numerous pecan orchards in Georgia. In some instances, 60% of the trees were diseased. Galled trees were less vigorous than noninfected trees. Among the pathogenic Agrobacterium strains isolated from 18 galled trees from six counties, biovar 1 strains predominated and most were sensitive to agrocin 84 in vitro. A representative biovar 1 strain from pecan was inhibited from infecting tomato seedlings by A. radiobacter K84. We would expect that biological control of crown gallin Georgia pecan orchards with strain K84 would be successful. Because the taxonomy of the members of the Rhizobiaceae has not been resolved by classical and modern methods of microbial classification, a serological analysis of the 50S ribosomal subunits was made. Antisera raised against 50S ribosomal subunits of five different agrobacteria were used to arrange 31 Agrobacterium strains and 6 Rhizobium strains into 16 serovars. The immunological relationships of 50S subunits of Agrobacterium did not confirm either of the major taxonomic groupings; fast-growing rhizobiacould not be immunologically differenciated from the agrobacteria andwere closely related to the biovar 3 strain CG64. The five antisera were specific at the family level. The antigenic structure of 50S ribosomal subunits of tumorigenic strain C58 and rhizogenic strain A4 were compared to that of their avirulent derivatives (NT1 and A4R1, respectively) and to strain A323 (A NT1 that was transformed with the large nopaline plasmid from K84) using Ouchterlony doubly diffusion tests. -
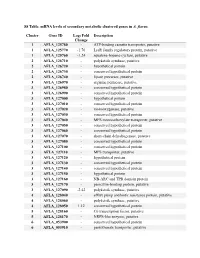
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

Comparative Genomics of Bradyrhizobium Japonicum CPAC
Siqueira et al. BMC Genomics 2014, 15:420 http://www.biomedcentral.com/1471-2164/15/420 RESEARCH ARTICLE Open Access Comparative genomics of Bradyrhizobium japonicum CPAC 15 and Bradyrhizobium diazoefficiens CPAC 7: elite model strains for understanding symbiotic performance with soybean Arthur Fernandes Siqueira1,2†, Ernesto Ormeño-Orrillo3†,RangelCelsoSouza4,ElisetePainsRodrigues5, Luiz Gonzaga Paula Almeida4, Fernando Gomes Barcellos5, Jesiane Stefânia Silva Batista6, Andre Shigueyoshi Nakatani2, Esperanza Martínez-Romero3, Ana Tereza Ribeiro Vasconcelos4 and Mariangela Hungria1,2* Abstract Background: The soybean-Bradyrhizobium symbiosis can be highly efficient in fixing nitrogen, but few genomic sequences of elite inoculant strains are available. Here we contribute with information on the genomes of two commercial strains that are broadly applied to soybean crops in the tropics. B. japonicum CPAC 15 (=SEMIA 5079) is outstanding in its saprophytic capacity and competitiveness, whereas B. diazoefficiens CPAC 7 (=SEMIA 5080) is known for its high efficiency in fixing nitrogen. Both are well adapted to tropical soils. The genomes of CPAC 15 and CPAC 7 were compared to each other and also to those of B. japonicum USDA 6T and B. diazoefficiens USDA 110T. Results: Differences in genome size were found between species, with B. japonicum having larger genomes than B. diazoefficiens. Although most of the four genomes were syntenic, genome rearrangements within and between species were observed, including events in the symbiosis island. In addition to the symbiotic region, several genomic islands were identified. Altogether, these features must confer high genomic plasticity that might explain adaptation and differences in symbiotic performance. It was not possible to attribute known functions to half of the predicted genes. -

Actes Congrès MICROBIOD 1
Actes du congrès international "Biotechnologie microbienne au service du développement" (MICROBIOD) Marrakech, MAROC 02-05 Novembre 2009 "Ce travail est publié avec le soutien du Ministère de l'Education Nationale, de l'Enseignement Supérieur, de la Formation des Cadres et de la Recherche Scientifique et du CNRST". MICROBIONA Edition www.ucam.ac.ma/microbiona 1 1 Mot du Comité d'organisation Cher (e) membre de MICROBIONA Cher(e) Participant(e), Sous le Haut Patronage de sa Majesté le Roi Mohammed VI, l'Association MICROBIONA et la Faculté des Sciences Semlalia, Université Cadi Ayyad, organisent du 02 au 05 Novembre 2009, en collaboration avec la Société Française de Microbiologie, le Pôle de Compétences Eau et Environnement et l'Incubateur Universitaire de Marrakech, le congrès international "Biotechnologie microbienne au service du développement" (MICROBIOD). Cette manifestation scientifique spécialisée permettra la rencontre de chercheurs de renommée internationale dans le domaine de la biotechnologie microbienne. Le congrès MICROBIOD est une opportunité pour les participants de mettre en relief l’importance socio-économique et environnementale de la valorisation et l’application de nouvelles techniques de biotechnologie microbienne dans divers domaines appliquées au développement et la gestion durable des ressources, à savoir l’agriculture, l'alimentation, l'environnement, la santé, l’industrie agro-alimentaire et le traitement et recyclage des eaux et des déchets par voie microbienne. Ce congrès sera également l’occasion pour les enseignants-chercheurs et les étudiants-chercheurs marocains d’actualiser leurs connaissances dans le domaine des biotechnologies microbiennes, qui serviront à l’amélioration de leurs recherches et enseignements. Le congrès MICROBIOD sera également l’occasion pour certains de nos collaborateurs étrangers de contribuer de près à la formation de nos étudiants en biotechnologie microbienne et ceci par la discussion des protocoles de travail, des méthodes d’analyse et des résultats obtenus. -

Revised Taxonomy of the Family Rhizobiaceae, and Phylogeny of Mesorhizobia Nodulating Glycyrrhiza Spp
Division of Microbiology and Biotechnology Department of Food and Environmental Sciences University of Helsinki Finland Revised taxonomy of the family Rhizobiaceae, and phylogeny of mesorhizobia nodulating Glycyrrhiza spp. Seyed Abdollah Mousavi Academic Dissertation To be presented, with the permission of the Faculty of Agriculture and Forestry of the University of Helsinki, for public examination in lecture hall 3, Viikki building B, Latokartanonkaari 7, on the 20th of May 2016, at 12 o’clock noon. Helsinki 2016 Supervisor: Professor Kristina Lindström Department of Environmental Sciences University of Helsinki, Finland Pre-examiners: Professor Jaakko Hyvönen Department of Biosciences University of Helsinki, Finland Associate Professor Chang Fu Tian State Key Laboratory of Agrobiotechnology College of Biological Sciences China Agricultural University, China Opponent: Professor J. Peter W. Young Department of Biology University of York, England Cover photo by Kristina Lindström Dissertationes Schola Doctoralis Scientiae Circumiectalis, Alimentariae, Biologicae ISSN 2342-5423 (print) ISSN 2342-5431 (online) ISBN 978-951-51-2111-0 (paperback) ISBN 978-951-51-2112-7 (PDF) Electronic version available at http://ethesis.helsinki.fi/ Unigrafia Helsinki 2016 2 ABSTRACT Studies of the taxonomy of bacteria were initiated in the last quarter of the 19th century when bacteria were classified in six genera placed in four tribes based on their morphological appearance. Since then the taxonomy of bacteria has been revolutionized several times. At present, 30 phyla belong to the domain “Bacteria”, which includes over 9600 species. Unlike many eukaryotes, bacteria lack complex morphological characters and practically phylogenetically informative fossils. It is partly due to these reasons that bacterial taxonomy is complicated. -

Defining the Rhizobium Leguminosarum Species Complex
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 December 2020 doi:10.20944/preprints202012.0297.v1 Article Defining the Rhizobium leguminosarum species complex J. Peter W. Young 1,*, Sara Moeskjær 2, Alexey Afonin 3, Praveen Rahi 4, Marta Maluk 5, Euan K. James 5, Maria Izabel A. Cavassim 6, M. Harun-or Rashid 7, Aregu Amsalu Aserse 8, Benjamin J. Perry 9, En Tao Wang 10, Encarna Velázquez 11, Evgeny E. Andronov 12, Anastasia Tampakaki 13, José David Flores Félix 14, Raúl Rivas González 11, Sameh H. Youseif 15, Marc Lepetit 16, Stéphane Boivin 16, Beatriz Jorrin 17, Gregory J. Kenicer 18, Álvaro Peix 19, Michael F. Hynes 20, Martha Helena Ramírez-Bahena 21, Arvind Gulati 22 and Chang-Fu Tian 23 1 Department of Biology, University of York, York YO10 5DD, UK 2 Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark; [email protected] 3 Laboratory for genetics of plant-microbe interactions, ARRIAM, Pushkin, 196608 Saint-Petersburg, Russia; [email protected] 4 National Centre for Microbial Resource, National Centre for Cell Science, Pune, India; [email protected] 5 Ecological Sciences, The James Hutton Institute, Invergowrie, Dundee DD2 5DA, UK; [email protected] (M.M.); [email protected] (E.K.J.) 6 Department of Ecology and Evolutionary Biology, University of California, Los Angeles, CA 90095, USA; [email protected] 7 Biotechnology Division, Bangladesh Institute of Nuclear Agriculture (BINA), Bangladesh; [email protected] 8 Ecosystems and Environment Research programme , Faculty of Biological and Environmental Sciences, University of Helsinki, FI-00014 Finland; [email protected] 9 Department of Microbiology and Immunology, University of Otago, Dunedin 9016, New Zealand; [email protected] 10 Departamento de Microbiología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Cd. -

Taxonomy and Systematics of Plant Probiotic Bacteria in the Genomic Era
AIMS Microbiology, 3(3): 383-412. DOI: 10.3934/microbiol.2017.3.383 Received: 03 March 2017 Accepted: 22 May 2017 Published: 31 May 2017 http://www.aimspress.com/journal/microbiology Review Taxonomy and systematics of plant probiotic bacteria in the genomic era Lorena Carro * and Imen Nouioui School of Biology, Newcastle University, Newcastle upon Tyne, UK * Correspondence: Email: [email protected]. Abstract: Recent decades have predicted significant changes within our concept of plant endophytes, from only a small number specific microorganisms being able to colonize plant tissues, to whole communities that live and interact with their hosts and each other. Many of these microorganisms are responsible for health status of the plant, and have become known in recent years as plant probiotics. Contrary to human probiotics, they belong to many different phyla and have usually had each genus analysed independently, which has resulted in lack of a complete taxonomic analysis as a group. This review scrutinizes the plant probiotic concept, and the taxonomic status of plant probiotic bacteria, based on both traditional and more recent approaches. Phylogenomic studies and genes with implications in plant-beneficial effects are discussed. This report covers some representative probiotic bacteria of the phylum Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes, but also includes minor representatives and less studied groups within these phyla which have been identified as plant probiotics. Keywords: phylogeny; plant; probiotic; PGPR; IAA; ACC; genome; metagenomics Abbreviations: ACC 1-aminocyclopropane-1-carboxylate ANI average nucleotide identity FAO Food and Agriculture Organization DDH DNA-DNA hybridization IAA indol acetic acid JA jasmonic acid OTUs Operational taxonomic units NGS next generation sequencing PGP plant growth promoters WHO World Health Organization PGPR plant growth-promoting rhizobacteria 384 1. -

Specificity in Legume-Rhizobia Symbioses
International Journal of Molecular Sciences Review Specificity in Legume-Rhizobia Symbioses Mitchell Andrews * and Morag E. Andrews Faculty of Agriculture and Life Sciences, Lincoln University, PO Box 84, Lincoln 7647, New Zealand; [email protected] * Correspondence: [email protected]; Tel.: +64-3-423-0692 Academic Editors: Peter M. Gresshoff and Brett Ferguson Received: 12 February 2017; Accepted: 21 March 2017; Published: 26 March 2017 Abstract: Most species in the Leguminosae (legume family) can fix atmospheric nitrogen (N2) via symbiotic bacteria (rhizobia) in root nodules. Here, the literature on legume-rhizobia symbioses in field soils was reviewed and genotypically characterised rhizobia related to the taxonomy of the legumes from which they were isolated. The Leguminosae was divided into three sub-families, the Caesalpinioideae, Mimosoideae and Papilionoideae. Bradyrhizobium spp. were the exclusive rhizobial symbionts of species in the Caesalpinioideae, but data are limited. Generally, a range of rhizobia genera nodulated legume species across the two Mimosoideae tribes Ingeae and Mimoseae, but Mimosa spp. show specificity towards Burkholderia in central and southern Brazil, Rhizobium/Ensifer in central Mexico and Cupriavidus in southern Uruguay. These specific symbioses are likely to be at least in part related to the relative occurrence of the potential symbionts in soils of the different regions. Generally, Papilionoideae species were promiscuous in relation to rhizobial symbionts, but specificity for rhizobial genus appears to hold at the tribe level for the Fabeae (Rhizobium), the genus level for Cytisus (Bradyrhizobium), Lupinus (Bradyrhizobium) and the New Zealand native Sophora spp. (Mesorhizobium) and species level for Cicer arietinum (Mesorhizobium), Listia bainesii (Methylobacterium) and Listia angolensis (Microvirga). -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341