FRAC Code List ©*2018: Fungicides Sorted by Mode of Action (Including FRAC Code Numbering)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
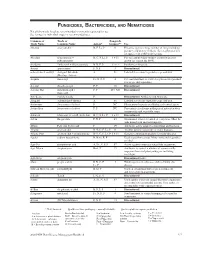
Fungicides, Bactericides, and Nematicides Not All Chemicals Listed Are Recommended Or Currently Registered for Use
FUNGICIDES, BACTERICIDES, AND NEMATICIDES Not all chemicals listed are recommended or currently registered for use. See listings for individual crops for recommended uses. Common or Trade or Fungicide Trade Name Common Name Action* Group #** Use Abound azoxystrobin B, F, Ls, P 11 Effective against a large number of fungi including powdery and downy mildews. Severe phytotoxicity on apples with a McIntosh heritage. Absolute tebuconazole + B, C, F, Ls, P 3 + 11 For rust and powdery mildew control in grasses trifloxystrobin grown for seed in the PNW. Academy fludioxonil + difenoconazole B-N, F, P 12 + 3 Postharvest fungicide. Accrue spiroxamine F, N, P 5 Discontinued. acibenzolar-S-methyl Actigard, Blockade A P1 Labeled for certain vegetable crops and fruit. (Heritage Action) Acquire metalaxyl Fs, N, P, S 4 For seed treatment to control ooymcetes in specified row crops and vegetables. Acrobat dimethomorph F, P 40 Discontinued. Acrobat MZ dimethomorph + F, P 40 + M3 Discontinued. mancozeb Acti-dione cycloheximide F Discontinued. Antibiotic and fungicide. Actigard acibenzolar-S-methyl A P1 Labeled for certain vegetable crops and fruit. Actinovate Streptomyces lydicus F NC Filamentous bacteria as a Biological control agent. Actino-Iron Streptomyces lydicus F, P NC For control of soilborne pathogens of indoor/outdoor ornamentals and vegetable crops. Adament tebuconazole + trifloxystrobin B, C, F, Ls, P 3 + 11 Discontinued. Adorn fluopicolide F, N, P 43 Ornamental label for control of oomycetes. Must be tank-mixed with another fungicide. Affirm Polyoxin D zinc salt F 19 Antibiotic active against certain fungi and bacteria. Aframe azoxystrobin B-N, C, F, Ls, P 11 Another generic fungicide for many diseases. -
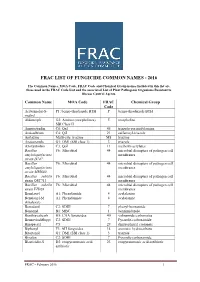
Frac List of Fungicide Common Names - 2016
FRAC LIST OF FUNGICIDE COMMON NAMES - 2016 The Common Names, MOA Code, FRAC Code and Chemical Group names included in this list are those used in the FRAC Code List and the associated List of Plant Pathogenic Organisms Resistant to Disease Control Agents. Common Name MOA Code FRAC Chemical Group Code Acibenzolar-S- P1: benzo-thiadiazole BTH P benzo-thiadiazole BTH methyl Aldimorph G2: Amines (morpholines) 5 morpholine SBI Class II Ametoctradin C8: QxI 45 triazolo-pyrimidylamine Amisulbrom C4: QiI 21 sulfamoyl-triazole Anilazine Multi-site: triazine M8 triazine Azaconazole G1: DMI (SBI class 1) 3 triazole Azoxystrobin C3: QoI 11 methoxy-acrylates Bacillus F6: Microbial 44 microbial disrupters of pathogen cell amyloliquefaciens membranes strain D747 Bacillus F6: Microbial 44 microbial disrupters of pathogen cell amyloliquefaciens membranes strain MBI600 Bacillus subtilis F6: Microbial 44 microbial disrupters of pathogen cell strain QST713 membranes Bacillus subtilis F6: Microbial 44 microbial disrupters of pathogen cell strain FZB24 membranes Benalaxyl A1: Phenylamide 4 acylalanine Benalaxyl-M A1: Phenylamide 4 acylalanine (kiralaxyl) Benodanil C2: SDHI 7 phenyl-benzamide Benomyl B1: MBC 1 benzimidazole Benthiavalicarb H5: CAA fungicides 40 valinamide carbamates Benzovindiflupyr C2: SDHI 7 Pyrazole-carboxamide Binapacryl C5 29 dinitrophenyl crotonate Biphenyl F3: AH fungicides 14 aromatic hydrocarbons Bitertanol G1: DMI (SBI class 1) 3 triazole Bixafen C2: SDHI 7 Pyrazole-carboxamide Blasticidin-S D2: enopyranuronic acid 23 enopyranuronic -

Abc Transporters of the Wheat Pathogen Mycosphaerella Graminicola
ABC TRANSPORTERS OF THE WHEAT PATHOGEN MYCOSPHAERELLA GRAMINICOLA Lute-Harm Zwiers CENTRALE LANDBOUWCATALOGUS 0000 0911 2273 Promoter: Prof.dr .ir .P.J.G.M .d eWi t Hoogleraar Fytopathologie, inhe t bijzonder plant-pathogeen interacties Co-promotor: Dr. ir. M.A. de Waard Universitair Hoofddocent, Laboratorium voor Fytopathologie Promotiecommissie:Prof .Dr . U. Gisi, Syngenta Crop Protection, Switzerland Dr. G.H.J.Kema , Plant Research International Prof.Dr . M.R. Muller, Wageningen Universiteit Prof.Dr . L.H.W. van der Plas, Wageningen Universiteit ( '• / Lute-HarmZwier s ABC transporters of the wheat pathogen Mycosphaerella graminicola Proefschrift terverkrijgin g van degraa dva n doctor opgeza gva n derecto r magnificus van Wageningen Universiteit, Prof.dr . ir. L. Speelman, inhe t openbaar te verdedigen opwoensda g 1 mei200 2 desnamiddag st e vier uur in deAul a / ISBN 90-5808-613-5 Aan mijn opa, Lute Zwiers VoorKarin, Merit en Eline Contents Chapter1 General introduction and Outline Chapter2 Characterisation of the ABC transporter genes MgAtrl and 27 MgAtr2fro m thewhea t pathogen Mycosphaerella graminicola Chapter3 Efficient Agrobacterium tumefaciens-mediated gene disruption in 45 the phytopathogen Mycosphaerella graminicola Chapter4 ABC transporters of the wheat pathogen Mycosphaerella 59 graminicolaan d virulence Chapter 5 ABC transporters of Mycosphaerella graminicola function as 71 protectants against biotic and xenobiotic toxic compounds Chapter 6 The role of ABC-transporters in azole-resistant laboratory 85 mutants ofth ewhea t pathogen Mycosphaerellagraminicola Chapter7 General discussion 100 Summary 115 Samenvatting 119 Curriculum Vitae 123 Nawoord 125 CHAPTER 1 General Introduction and Outline Part of this Chapter has been published by A.C. Andrade, L-H. -
Availability of Substances in Europe Updated in November 2020 Available Substances in EU Under Reg 1107/2009
Availability of Substances in Europe Updated in November 2020 Available substances in EU under reg 1107/2009 Not PPP 15 Approved 475 Not approved Pending 902 42 Approved Pending Not approved Not PPP (17-11-20 / EU Pesticide Database) Updated on 17 /11 /2020 Status of approved substances Not in AIR programmes Renewed 15 substances renewed as (expiration > 2025) Candidates for Substitution (CfS) 73 60 Substance Function of which : of which : Alpha-Cypermethrin 15 on-going in a new AIR 38 new substances approved under the 1107/2009: (alphamethrin) Insecticide program - 16 substances of chemical origin Bordeaux mixture Fungicide - 22 substances non chemical (of which 14 low risk): Copper compounds Fungicide fungicide, bactericide, etc. Will be included in AIR Programme Copper hydroxide Fungicide Copper oxide Fungicide 22 basic substances (without expiration date) Copper oxychloride Fungicide Total Esfenvalerate Insecticide 475 Imazamox Herbicide approved Renewal on-going lambda-Cyhalothrin Insecticide Methoxyfenozide Insecticide 357 Metsulfuron-methyl Herbicide of which : Pendimethalin Herbicide 24 not supported Propyzamide Herbicide 56 candidates for substitution(CfS) Prosulfuron Herbicide Tribasic copper sulfate Fungicide 6 substances cut–off (R1) Updated on 17 /11 /2020 Evolution of availability of substances in Europe Evolution of availability of substances in Europe 600 90 85 556 80 500 494 492 485 73 477 479 475 468 466 70 64 64 400 60 60 58 55 55 352 53 54 52 51 51 49 50 300 45 45 40 34 200 30 27 Number substances ofactive Number of -

UC Riverside UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title Population Structure of the Sour Rot Pathogens Galactomyces citri-aurantii and G. geotrichum and Evaluation of Sterol Demethylation Inhibitors for Postharvest Management of Citrus Decays Permalink https://escholarship.org/uc/item/2nv0s0cg Author McKay, Alistair Hartley Publication Date 2011 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE Population Structure of the Sour Rot Pathogens Galactomyces citri-aurantii and G. geotrichum and Evaluation of Sterol Demethylation Inhibitors for Postharvest Management of Citrus Decays A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Plant Pathology by Alistair Hartley McKay March 2011 Dissertation Committee: Dr. James E. Adaskaveg, Chairperson Dr. Michael D. Coffey Dr. Michael E. Stanghellini Copyright by Alistair Hartley McKay 2011 The Dissertation of Alistair McKay is approved: __________________________________________________ __________________________________________________ __________________________________________________ Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I would like to thank the California citrus research board for continuing to support citrus research in California, and in particular this postharvest project which funded much of my research. I would like to acknowledge the support demonstrated by the developers of the postharvest chemistries we use to manage postharvest diseases of citrus including, Syngenta Crop Protection, Bayer CropScience, Dow AgroSciences, Janssen Pharmaceutica, JBT FoodTech. For assistance with residue analysis, formulations and coatings I would like to thank the staff at Decco US, Post- Harvest including Mohsen Sales, Sukas Wartanessian, Sonyia Villegas, Tommy Walia, and Mike Low. For the majority of my graduate program I was employed on a part-time basis with Sunkist Growers Inc. -

SOYBEAN DISEASE CONTROL John D
South Carolina Pest Management Handbook for Field Crops - 2017 SOYBEAN DISEASE CONTROL John D. Mueller, Extension Soybean Pathologist Soybeans can be affected by diseases throughout the growing season. In general seedling diseases are only a problem in fields planted very early in the growing season when soil temperatures are low. They are especially prevalent if low soil temperatures are combined with very wet soils. Usually by mid‐May seedling diseases are no longer a problem. Seed treatment fungicides normally do a good job of controlling seedling diseases. During mid‐season many leaf diseases are common on soybean. Downy Mildew, Brown Spot (Septoria Blight), Cercospora Leaf Spot and Frogeye Leaf Spot are very common and stem diseases such as Pod & Stem Blight and Anthracnose are common from midseason to harvest in wet years. Soybean Rust is active in South Carolina primarily after mid‐August in most years. Soybean varieties vary greatly in their susceptibility to diseases such as Frogeye Leaf Spot. Choosing a resistant variety is much more cost effective than fungicide applications. Fungicides are available that can help with many of these diseases. It is important to accurately identify the diseases you are trying to control as not all diseases can be controlled by all fungicides. Check the label of individual fungicides or your South Carolina Soybean Production Guide for information on accurate identification of diseases based on field symptoms. In general South Carolina soybeans should not be sprayed for disease control until after they flower. This is true whether Soybean Rust or other diseases are the target. Wet weather increases the severity of most fungal diseases and the subsequent need for a fungicide. -

Cyproconazole
766 Cyproconazole CYPROCONAZOLE (239) First draft prepared by Mr S Funk, United States Environmental Protection Agency, Washington DC, USA EXPLANATION Cyproconazole is an azole fungicide used to control a wide range of fungi on cereal crops, coffee, sugar beet, fruit trees, grapes, including rust on cereal crops, powdery mildew on cereal crops, fruit tree and grapes, and scab on apple. It is both a prevention and treatment fungicide. At the 41st session of the CCPR (2009), it was scheduled for the evaluation as a new compound by the 2010 JMPR. The residue studies were submitted by the manufacturer for support of the following commodities: almond, apple, barley, bean, maize, oat, pea, peanut, rice, sugar beet, soya bean, and wheat. GAP information was also submitted by Japan and the Netherlands. IDENTITY ISO Common Name Cyproconazole Chemical name: IUPAC (2RS,3RS;2SR,3SR)-2-(4-chlorophenyl)-3-cyclopropyl-1-(1H-1,2,4- triazol-1-yl)butan-2-ol CA alpha-(4-chlorophenyl)-alpha-(1-cyclopropyl-ethyl)-1H-1,2,4-triazole-1- ethanol CAS No 94361-06-5 Manufacturers code No. SAN619 Structural formula N N OH N Cl Molecular formula C15H18ClN3O Molecular mass 291.8 Stereochemistry: Cyproconazole possesses two chiral carbon atoms (C2 and C3 of the butane backbone) indicated in the structure with asterisks: Cyproconazole 767 * N N * N OH Cl This structure exists in four stereoisomeric forms: two enantiomeric pairs of diastereoisomers. The following are defined: “Diastereomer A”: enantiomeric pair, where the 3-hydroxy group and 2-hydrogen are located on the same side. “Diastereomer B”: enantiomeric pair, where the 3-hydroxy group and 2-hydrogen are located on opposite sides. -

Fungicide, Nematicide and Bactericide Table
Fung/Nemat/Bact Table Rev. 3/15 Fungicide, Nematicide and Bactericide Table The following information will help you interpret the Fungicide, Nematicide, and Bactericide Table: Restricted or Cancelled Pesticides Certain formulations, uses, or amounts of the pesticide compound have been restricted (R) or canceled (C). Check a current label (the label of the product to be purchased) to determine which are restricted or general uses. Pesticides or pesticide uses which are restricted require the applicator to be certified or working under supervision of a certified applicator. Common Names USDA, and later EPA, has assigned most pesticide chemicals an official common name. This name has been listed where possible. It should not be capitalized. Other Names Fungicides, bactericides, and nematicides originate from a wide range of compounds and are classified according to use: fungicide (F), nematicide (N), or bactericide (B). Toxicity Category and LD50 Values When pesticides are registered, the Environmental Protection Agency uses the acute LD50 values to determine the toxicity category and the words or symbols which must be placed on the label. The categories assigned in the following table are based on information available and may not reflect EPA’s toxicity category. LD50 is the dosage at which one-half of the test animals are killed. Usually rats are tested, although mice or rabbits may be used. LD50 is measured in milligrams of chemical being tested per kilogram of animal (mg/kg). One part per million (ppm) is equal to one mg/kg. LD50 is usually determined for the technical material rather than the formulated product. The higher the LD50, the less toxic the material is. -
2020 Fungicide Classification Chart
FUNGICIDE CLASSIFICATION Repeated use of fungicides with the same mode of action can result in the selection of fungicide-resistant strains of plant pathogens. by PREMIX This section lists premix fungicides by their trade by MODE OF ACTION (MOA) names so you can identify the premix’s component This section groups fungicides by their mode of action to assist in the selection of fungicides 1) to maintain greater fungicides and their respective mode of action groups. diversity in fungicide use and 2) to rotate among effective fungicides with different modes of action to delay the development Refer to the Mode of Action section on the left of fungicide resistance. for more information. ACTIVE FRAC FRAC MODE OF ACTION CHEMICAL ACTIVE PRODUCT EXAMPLES PREMIX CODE FAMILY INGREDIENT (Trade Name) INGREDIENT CODE MITOSIS DISRUPTERS thiophanate-methyl 1 ACROPOLIS tetraconazole 3 MBC (methyl benzimidazole carbamates) Topsin, multiple generics 1 Thiophanates thiophanate-methyl B1: ß-tubuline assembly in mitosis and component in premix tetraconazole 3 AFFIANCE azoxystrobin 11 CELL MEMBRANE DISRUPTERS propiconazole 3 AFRAME PLUS cyproconazole Alto and component in premix azoxystrobin 11 cyproconazole 3 Component of Quadris Top difenoconazole APROACH PRIMA and Miravis Top picoxystrobin 11 propiconazole 3 flutriafol Topguard and component in premix AVARIS azoxystrobin 11 Revysol, component of Revytek, mefentrifluconazole 3 Veltyma tebuconazole DMI (demethylation inhibitors)/ AZOXY TEB Triazoles azoxystrobin 11 triazoles metconazole Component of Headline -
![Profiling the Toxcast Library with a Pluripotent Human (H9) Stem Cell Line-Based Biomarker Assay for Developmental Toxici- Ty[AQ2][AQ3]](https://docslib.b-cdn.net/cover/0950/profiling-the-toxcast-library-with-a-pluripotent-human-h9-stem-cell-line-based-biomarker-assay-for-developmental-toxici-ty-aq2-aq3-7970950.webp)
Profiling the Toxcast Library with a Pluripotent Human (H9) Stem Cell Line-Based Biomarker Assay for Developmental Toxici- Ty[AQ2][AQ3]
1 Profiling the ToxCast Library With a Pluripotent Human (H9) Stem Cell Line-Based Biomarker Assay for Developmental Toxici- ty[AQ2][AQ3] PROFILING THE TOXCAST LIBRARY[AQ1] ZURLINDEN et al. Todd J. Zurlinden * Katerine S. Saili * Nathaniel Rush * Parth Kothiya * Richard S. Judson * Keith A. Houck * E. Sidney Hunter † Nancy C. Baker ‡ Jessica A. Palmer § Russell S. Thomas * and Thomas B. Knudsen*,1[AQ5][AQ4] *. National Center for Computational Toxicology (NCCT) and †. National Health and Environmental Effects Research Laboratory (NHEERL), Office of Research and Development (ORD), U.S. En- vironmental Protection Agency (USEPA), Research Triangle Park, North Carolina 27711; ‡. Leidos, Research Triangle Park, North Carolina 27711; and §. Stemina Biomarker Discovery, Inc, Madison, Wisconsin 53719[AQ6] 1. To whom correspondence should be addressed at National Center for Computational Toxicology (B205-01), U.S. Environmental Protection Agency, Research Triangle Park, NC 27711. E-mail: [email protected].[AQ7] Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not con‐ stitute endorsement or recommendation for use. ABSTRACT The Stemina devTOX quickPredict platform is a human pluripotent stem cell-based assay that predicts the developmental toxicity potential based on changes in cellular metabolism following chemical exposure [Palmer, J. A., Smith, A. M., Egnash, L. A., Conard, K. R., West, P. R., Burrier, R. E., Donley, E. L. R., and Kirchner, F. R. (2013). Establishment and assessment of a new human embry- onic stem cell-based biomarker assay for developmental toxicity screening. -

By PREMIX by MODE of ACTION
W 359 by PREMIX This section lists premix fungicides alphabetically by their trade names so you can identify the premix’s component fungicides and their respective site of action groups. Refer to the Site of Action section on the left for more information. ACTIVE FRAC PREMIX INGREDIENT (%) GROUP REPEATED USE OF FUNGICIDES WITH THE SAME SITE OF ACTION CAN RESULT Azoxystrobin 18.2% 11 IN THE DEVELOPMENT OF FUNGICIDE-RESISTANT PATHOGEN POPULATION. QUADRIS TOP Difenoconazole 11.4% 3 Azoxystrobin 7.0% 11 AVARIS Propiconazole 11.7% 3 Azoxystrobin 7.0% 11 QUILT Propiconazole 11.7% 3 Azoxystrobin 13.5% 11 QUILT XCEL by MODE OF ACTION Propiconazole 11.7% 3 (effect on plant pathogen) Picoxystrobin 17.94% 11 APROACH PRIMA This section groups fungicides by their modes of action to assist you in selecting fungicides 1) to maintain greater Cyproconazole 7.17% 3 diversity in fungicide use and 2) to rotate among effective fungicides with different sites of action to delay the Fluoxastrobin 18.0% 11 EVITO T development of fungicide resistance. Tebuconazole 25.0% 3 Fluoxastrobin 14.84% 11 FORTIX Flutriafol 19.3% 3 Pyraclostrobin 28.58% 11 PRIAXOR FRAC SITE OF ACTION CHEMICAL ACTIVE PRODUCT EXAMPLES Fluxapyroxad 14.33% 7 CODE FAMILY INGREDIENT (TRADE NAME) Pyraclostrobin 28.58% 11 MITOSIS PRIAXOR D Fluxapyroxad 14.33% 7 DISRUPTERS MBC (Methyl Benzimidazole Carbamates) Topsin, multiple generics and Tetraconazole 20.5% 3 Thiophanates Thiophanate-methyl 1 B1: ß-tubuline assembly in mitosis component in premix Trifloxystrobin 11.4% 11 CELL MEMBRANE STRATEGO -

Conazole Fungicides Inhibit Leydig Cell Testosterone Secretion And
Toxicology Reports 1 (2014) 271–283 View metadata, citation and similar papers at core.ac.uk brought to you by CORE Contents lists available at ScienceDirect provided by Elsevier - Publisher Connector Toxicology Reports journa l homepage: www.elsevier.com/locate/toxrep Conazole fungicides inhibit Leydig cell testosterone secretion and androgen receptor activation in vitro a,b,∗ a b,a Maarke J.E. Roelofs , A. Roberto Temming , Aldert H. Piersma , a a Martin van den Berg , Majorie B.M. van Duursen a Endocrine Toxicology Research Group, Institute for Risk Assessment Sciences (IRAS), Utrecht University, P.O. Box 80.177, NL-3508 TD Utrecht, The Netherlands b Center for Health Protection, National Institute for Public Health and the Environment (RIVM), P.O. Box 1, 3720 BA Bilthoven, The Netherlands a r t i c l e i n f o a b s t r a c t Article history: Conazole fungicides are widely used in agriculture despite their suspected endocrine dis- Received 24 March 2014 rupting properties. In this study, the potential (anti-)androgenic effects of ten conazoles Received in revised form 12 May 2014 were assessed and mutually compared with existing data. Effects of cyproconazole (CYPRO), Accepted 12 May 2014 fluconazole (FLUC), flusilazole (FLUS), hexaconazole (HEXA), myconazole (MYC), pencona- Available online 22 May 2014 zole (PEN), prochloraz (PRO), tebuconazole (TEBU), triadimefon (TRIA), and triticonazole (TRIT) were examined using murine Leydig (MA-10) cells and human T47D-ARE cells sta- Chemical compounds studied in this article: bly transfected with an androgen responsive element and a firefly luciferase reporter gene. Cyproconazole (PubChem CID: 86132) Six conazoles caused a decrease in basal testosterone (T) secretion by MA-10 cells varying Fluconazole (PubChem CID: 3365) from 61% up to 12% compared to vehicle-treated control.