Structural Evolution of Nrdna ITS in Pinaceae and Its Phylogenetic Implications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of Fir Species in the Silviculture of British Forests
Kastamonu Üni., Orman Fakültesi Dergisi, 2012, Özel Sayı: 15-26 Kastamonu Univ., Journal of Forestry Faculty, 2012, Special Issue The Role of True Fir Species in the Silviculture of British Forests: past, present and future W.L. MASON Forest Research, Northern Research Station, Roslin, Midlothian, Scotland EH25 9SY, U.K. E.mail:[email protected] Abstract There are no true fir species (Abies spp.) native to the British Isles: the first to be introduced was Abies alba in the 1600s which was planted on some scale until the late 1800s when it proved vulnerable to an insect pest. Thereafter interest switched to North American species, particularly grand (Abies grandis) and noble (Abies procera) firs. Provenance tests were established for A. alba, A. amabilis, A. grandis, and A. procera. Other silver fir species were trialled in forest plots with varying success. Although species such as grand fir have proved highly productive on favourable sites, their initial slow growth on new planting sites and limited tolerance of the moist nutrient-poor soils characteristic of upland Britain restricted their use in the afforestation programmes of the last century. As a consequence, in 2010, there were about 8000 ha of Abies species in Britain, comprising less than one per cent of the forest area. Recent species trials have confirmed that best growth is on mineral soils and that, in open ground conditions, establishment takes longer than for other conifers. However, changes in forest policies increasingly favour the use of Continuous Cover Forestry and the shade tolerant nature of many fir species makes them candidates for use with selection or shelterwood silvicultural systems. -
Download Validation Data
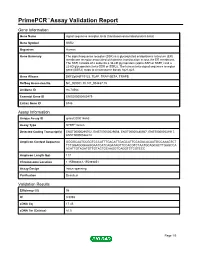
PrimePCR™Assay Validation Report Gene Information Gene Name signal sequence receptor, beta (translocon-associated protein beta) Gene Symbol SSR2 Organism Human Gene Summary The signal sequence receptor (SSR) is a glycosylated endoplasmic reticulum (ER) membrane receptor associated with protein translocation across the ER membrane. The SSR consists of 2 subunits a 34-kD glycoprotein (alpha-SSR or SSR1) and a 22-kD glycoprotein (beta-SSR or SSR2). The human beta-signal sequence receptor gene (SSR2) maps to chromosome bands 1q21-q23. Gene Aliases DKFZp686F19123, TLAP, TRAP-BETA, TRAPB RefSeq Accession No. NC_000001.10, NT_004487.19 UniGene ID Hs.74564 Ensembl Gene ID ENSG00000163479 Entrez Gene ID 6746 Assay Information Unique Assay ID qHsaCID0014663 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000295702, ENST00000529008, ENST00000480567, ENST00000531917, ENST00000526212 Amplicon Context Sequence GGGGCAATCCGGTCCCATTTGACATTGAGCATTCCAGACACAATGCCAAAGTCT TCTGGAGGGAAGGAATCATCAGATAGTTCCACGTCTAATGCAGCACTTGAGCCA ACATTGTAGATGTTGTACTGCAAGGTCAGGTCTCGTCCC Amplicon Length (bp) 117 Chromosome Location 1:155988061-155989851 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9998 cDNA Cq 17.45 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SSR2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

A Crosstalk Between the RNA Binding Protein Smaug and the Hedgehog Pathway Links Cell Signaling to Mrna Regulation in Drosophila Lucía Bruzzone
A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila Lucía Bruzzone To cite this version: Lucía Bruzzone. A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila. Cellular Biology. Université Sorbonne Paris Cité, 2018. English. NNT : 2018USPCC234. tel-02899776 HAL Id: tel-02899776 https://tel.archives-ouvertes.fr/tel-02899776 Submitted on 15 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparée à l’Université Paris Diderot Ecole doctorale HOB n° 561 Institut Jacques Monod / Equipe Développement, Signalisation et Trafic A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in Drosophila Lucía Bruzzone Thèse de doctorat de Biologie Dirigée par Anne Plessis Présentée et soutenue publiquement à Paris le 19 mars 2018 Président du jury: Alain Zider / Professeur Université Paris Diderot -

Disturbances Influence Trait Evolution in Pinus
Master's Thesis Diversify or specialize: Disturbances influence trait evolution in Pinus Supervision by: Prof. Dr. Elena Conti & Dr. Niklaus E. Zimmermann University of Zurich, Institute of Systematic Botany & Swiss Federal Research Institute WSL Birmensdorf Landscape Dynamics Bianca Saladin October 2013 Front page: Forest of Pinus taeda, northern Florida, 1/2013 Table of content 1 STRONG PHYLOGENETIC SIGNAL IN PINE TRAITS 5 1.1 ABSTRACT 5 1.2 INTRODUCTION 5 1.3 MATERIAL AND METHODS 8 1.3.1 PHYLOGENETIC INFERENCE 8 1.3.2 TRAIT DATA 9 1.3.3 PHYLOGENETIC SIGNAL 9 1.4 RESULTS 11 1.4.1 PHYLOGENETIC INFERENCE 11 1.4.2 PHYLOGENETIC SIGNAL 12 1.5 DISCUSSION 14 1.5.1 PHYLOGENETIC INFERENCE 14 1.5.2 PHYLOGENETIC SIGNAL 16 1.6 CONCLUSION 17 1.7 ACKNOWLEDGEMENTS 17 1.8 REFERENCES 19 2 THE ROLE OF FIRE IN TRIGGERING DIVERSIFICATION RATES IN PINE SPECIES 21 2.1 ABSTRACT 21 2.2 INTRODUCTION 21 2.3 MATERIAL AND METHODS 24 2.3.1 PHYLOGENETIC INFERENCE 24 2.3.2 DIVERSIFICATION RATE 24 2.4 RESULTS 25 2.4.1 PHYLOGENETIC INFERENCE 25 2.4.2 DIVERSIFICATION RATE 25 2.5 DISCUSSION 29 2.5.1 DIVERSIFICATION RATE IN RESPONSE TO FIRE ADAPTATIONS 29 2.5.2 DIVERSIFICATION RATE IN RESPONSE TO DISTURBANCE, STRESS AND PLEIOTROPIC COSTS 30 2.5.3 CRITICAL EVALUATION OF THE ANALYSIS PATHWAY 33 2.5.4 PHYLOGENETIC INFERENCE 34 2.6 CONCLUSIONS AND OUTLOOK 34 2.7 ACKNOWLEDGEMENTS 35 2.8 REFERENCES 36 3 SUPPLEMENTARY MATERIAL 39 3.1 S1 - ACCESSION NUMBERS OF GENE SEQUENCES 40 3.2 S2 - TRAIT DATABASE 44 3.3 S3 - SPECIES DISTRIBUTION MAPS 58 3.4 S4 - DISTRIBUTION OF TRAITS OVER PHYLOGENY 81 3.5 S5 - PHYLOGENETIC SIGNAL OF 19 BIOCLIM VARIABLES 84 3.6 S6 – COMPLETE LIST OF REFERENCES 85 2 Introduction to the Master's thesis The aim of my master's thesis was to assess trait and niche evolution in pines within a phylogenetic comparative framework. -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -

North American Pinyon-Juniper Woodlands: Ecological Composition, Dynamics, and Future Trends
North American Pinyon–Juniper Woodlands: Ecological Composition, Dynamics, and Future Trends Esteban Muldavin and F Jack Triepke, University of New Mexico, Albuquerque, NM, United States © 2019 Elsevier Inc. All rights reserved. Introduction 1 Types of Pinyon–Juniper Woodlands 3 Detailed Pinyon–Juniper Woodland Descriptions 4 G198 Californian Conifer Forest & Woodland Group 4 G487 Madrean Juniper Open Woodland Group 5 G200 Madrean Pinyon–Juniper Woodland Group 5 G900 Colorado Plateau Pinyon–Juniper Woodland Group 5 G248 Columbia Plateau Western Juniper Open Woodland Group 6 G899 Great Basin Pinyon–Juniper Woodland Group 6 G249 Intermountain Basins Curl-leaf Mountain-mahogany Woodland & Scrub 6 G252 Southern Rocky Mountain Juniper Open Woodland Group 6 G253 Southern Rocky Mountain Pinyon–Juniper Woodland Group 6 Ecological Dynamics in Pinyon–Juniper Woodlands 7 Fire Regimes 7 Insect Pathogens 8 Ecosystem Services and the Changing Land Use of Pinyon–Juniper Woodlands 9 History and Land Use Impacts (e.g., Involving Herbivory, Changes in Fire Regime) 10 Pinyon nuts 10 Livestock use 10 Fuelwood 11 Pinyon–Juniper Woodlands and Climate 12 Tree Mortality and Plant Composition Changes 12 Carbon Storage as an Ecosystem Service 13 Conclusion—Climate and Carbon 13 Future Trends and Conservation 14 References 14 Abstract Pinyon and juniper woodland is a major biome type centered on the Basin and Range and Colorado Plateau physiographic regions of interior western North America. It covers over 40 million ha from western Oregon in the northwest United States, eastward to Wyoming and southward to northern Chihuahua in Mexico, then westward to Baja California. Climatically, these woodlands lie within both the warm and cold temperate zones of North America but at the wetter and cooler portion of the semiarid realm where precipitation seldom exceeds 400 mm. -

California Partners in Flight the USDA Forest Service Klamath Bird Observatory and PRBO Conservation Science
The Coniferous Forest Bird Conservation Plan A Strategy for Protecting and Managing Coniferous Forest Habitats and Associated Birds in California Version 1.1 March 2002 A project of California Partners in Flight The USDA Forest Service Klamath Bird Observatory and PRBO Conservation Science Conservation Plan Lead Authors: John C. Robinson, USDA Forest Service John Alexander, Klamath Bird Observatory Conservation Plan Supporting Authors, PRBO Conservation Science: Sue Abbott Diana Humple Grant Ballard Melissa Pitkin Dan Barton Sandy Scoggin Gregg Elliott Diana Stralberg Sacha Heath Focal Species Account Authors: Black-backed Woodpecker – Kerry Farris Black-throated Gray Warbler – Tina Mark, USDA Forest Service Brown Creeper – Danielle LeFer, San Francisco Bay Bird Observatory Dark-eyed Junco – Jim DeStaebler, PRBO Conservation Science Flammulated Owl – Susan Yasuda, USDA Forest Service Fox Sparrow – Anne King, EDAW, Inc. Golden-crowned Kinglet – John C. Robinson, USDA Forest Service MacGillivray's Warbler – Chris Otahal, USDA Forest Service Olive-sided Flycatcher – Paul Brandy, Endangered Species Recovery Program Pileated Woodpecker – John C. Robinson, USDA Forest Service Red-breasted Nuthatch – Tina Mark and John C. Robinson, USDA Forest Service Vaux's Swift – John Sterling, Jones and Stokes Associates Western Tanager – Cory Davis, USDA Forest Service Financial Contributors: USDA Forest Service Packard Foundation National Fish and Wildlife Foundation PRBO Conservation Science Klamath Bird Observatory Acknowledgements: California Partners in Flight wishes to thank everyone who helped write, promote, and produce this document. Special thanks to Laurie Fenwood, Geoffrey Geupel, Aaron Holmes, Genny Wilson, Ryan Burnett, and Doug Wallace, and to Sophie Webb for her cover illustration. Recommended Citation: CalPIF (California Partners in Flight). 2002. -

Newly Identified Gon4l/Udu-Interacting Proteins
www.nature.com/scientificreports OPEN Newly identifed Gon4l/ Udu‑interacting proteins implicate novel functions Su‑Mei Tsai1, Kuo‑Chang Chu1 & Yun‑Jin Jiang1,2,3,4,5* Mutations of the Gon4l/udu gene in diferent organisms give rise to diverse phenotypes. Although the efects of Gon4l/Udu in transcriptional regulation have been demonstrated, they cannot solely explain the observed characteristics among species. To further understand the function of Gon4l/Udu, we used yeast two‑hybrid (Y2H) screening to identify interacting proteins in zebrafsh and mouse systems, confrmed the interactions by co‑immunoprecipitation assay, and found four novel Gon4l‑interacting proteins: BRCA1 associated protein‑1 (Bap1), DNA methyltransferase 1 (Dnmt1), Tho complex 1 (Thoc1, also known as Tho1 or HPR1), and Cryptochrome circadian regulator 3a (Cry3a). Furthermore, all known Gon4l/Udu‑interacting proteins—as found in this study, in previous reports, and in online resources—were investigated by Phenotype Enrichment Analysis. The most enriched phenotypes identifed include increased embryonic tissue cell apoptosis, embryonic lethality, increased T cell derived lymphoma incidence, decreased cell proliferation, chromosome instability, and abnormal dopamine level, characteristics that largely resemble those observed in reported Gon4l/udu mutant animals. Similar to the expression pattern of udu, those of bap1, dnmt1, thoc1, and cry3a are also found in the brain region and other tissues. Thus, these fndings indicate novel mechanisms of Gon4l/ Udu in regulating CpG methylation, histone expression/modifcation, DNA repair/genomic stability, and RNA binding/processing/export. Gon4l is a nuclear protein conserved among species. Animal models from invertebrates to vertebrates have shown that the protein Gon4-like (Gon4l) is essential for regulating cell proliferation and diferentiation. -

Wa Shan – Emei Shan, a Further Comparison
photograph © Zhang Lin A rare view of Wa Shan almost minus its shroud of mist, viewed from the Abies fabri forested slopes of Emei Shan. At its far left the mist-filled Dadu River gorge drops to 500-600m. To its right the 3048m high peak of Mao Kou Shan climbed by Ernest Wilson on 3 July 1903. “As seen from the top of Mount Omei, it resembles a huge Noah’s Ark, broadside on, perched high up amongst the clouds” (Wilson 1913, describing Wa Shan floating in the proverbial ‘sea of clouds’). Wa Shan – Emei Shan, a further comparison CHRIS CALLAGHAN of the Australian Bicentennial Arboretum 72 updates his woody plants comparison of Wa Shan and its sister mountain, World Heritage-listed Emei Shan, finding Wa Shan to be deserving of recognition as one of the planet’s top hotspots for biological diversity. The founding fathers of modern day botany in China all trained at western institutions in Europe and America during the early decades of last century. In particular, a number of these eminent Chinese botanists, Qian Songshu (Prof. S. S. Chien), Hu Xiansu (Dr H. H. Hu of Metasequoia fame), Chen Huanyong (Prof. W. Y. Chun, lead author of Cathaya argyrophylla), Zhong Xinxuan (Prof. H. H. Chung) and Prof. Yung Chen, undertook their training at various institutions at Harvard University between 1916 and 1926 before returning home to estab- lish the initial Chinese botanical research institutions, initiate botanical exploration and create the earliest botanical gardens of China (Li 1944). It is not too much to expect that at least some of them would have had personal encounters with Ernest ‘Chinese’ Wilson who was stationed at the Arnold Arboretum of Harvard between 1910 and 1930 for the final 20 years of his life. -

(FIA) Annual Inventory Answers the Question: What Is Happening to Pinyon-Juniper Woodlands?
Forest Inventory and Analysis (FIA) Annual Inventory Answers the Question: What Is Happening to Pinyon-Juniper Woodlands? John D. Shaw, Brytten E. Steed, and Larry T. DeBlander Widespread mortality in the pinyon-juniper forest type is associated with several years of drought in Pinus monophylla Torr.&Frem.)—and one the southwestern United States. A complex of drought, insects, and disease is responsible for pinyon or more juniper species (Juniperus spp.); mortality rates approaching 100% in some areas, while other areas have experienced little or no pure stands of pinyon usually are not con- mortality. Implementation of the Forest Inventory and Analysis (FIA) annual inventory in several states sidered a separate type. Pinyon nuts were a coincided with the onset of elevated mortality rates. Adjunct inventories provided supplemental data on staple food for Native Americans of the damaging agents. Preliminary analysis reveals the status and trends of mortality in pinyon-juniper Southwest for thousands of years, and the woodlands. harvest of this valuable resource continues ABSTRACT today (Lanner 1981). There also are many Keywords: forest inventory, FIA, pinyon pines, pinyon-juniper woodlands, drought, mortality, Ips traditional and modern uses for pinyon and confusus, bark beetles, southwestern United States juniper wood. The extensive range and vol- ume of the resource has generated interest in intensive use, such as an energy-producing biomass crop. However, pinyon-juniper he Forest Inventory and Analysis same systematic sample grid as was used for woodland is seen as a weedy invader of pro- (FIA) is a national USDA Forest periodic inventories, but the plots are evenly ductive grasslands by some. -

Case Report an Adolescent with a Rare De Novo Distal Trisomy 6P and Distal Monosomy 6Q Chromosomal Combination
Hindawi Case Reports in Genetics Volume 2020, Article ID 8857628, 8 pages https://doi.org/10.1155/2020/8857628 Case Report An Adolescent with a Rare De Novo Distal Trisomy 6p and Distal Monosomy 6q Chromosomal Combination Leia A. Peterman , Gail H. Vance, Erin E. Conboy, Katelynn Anderson, and David D. Weaver Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA Correspondence should be addressed to David D. Weaver; [email protected] Received 15 April 2020; Revised 11 August 2020; Accepted 23 August 2020; Published 31 August 2020 Academic Editor: Silvia Paracchini Copyright © 2020 Leia A. Peterman et al. (is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. We report on a 12-year-old female with both a partial duplication and deletion involving chromosome 6. (e duplication involves 6p25.3p24.3 (7.585 Mb) while the deletion includes 6q27q27 (6.244 Mb). (is chromosomal abnormality is also described as distal trisomy 6p and distal monosomy 6q. (e patient has a Chiari II malformation, hydrocephalus, agenesis of the corpus callosum, microcephaly, bilateral renal duplicated collecting system, scoliosis, and myelomeningocele associated with a neurogenic bladder and bladder reflux. Additional features have included seizures, feeding dysfunction, failure to thrive, sleep apnea, global de- velopmental delay, intellectual disability, and absent speech. To our knowledge, our report is just the sixth case in the literature with concomitant distal 6p duplication and distal 6q deletion. Although a majority of chromosomal duplication-deletion cases have resulted from a parental pericentric inversion, the parents of our case have normal chromosomes.