Structure of the Mammalian Oligosaccharyl-Transferase Complex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 14: Functional Genomics Learning Objectives
Chapter 14: Functional Genomics Learning objectives Upon reading this chapter, you should be able to: ■ define functional genomics; ■ describe the key features of eight model organisms; ■ explain techniques of forward and reverse genetics; ■ discuss the relation between the central dogma and functional genomics; and ■ describe proteomics-based approaches to functional genomics. Outline : Functional genomics Introduction Relation between genotype and phenotype Eight model organisms E. coli; yeast; Arabidopsis; C. elegans; Drosophila; zebrafish; mouse; human Functional genomics using reverse and forward genetics Reverse genetics: mouse knockouts; yeast; gene trapping; insertional mutatgenesis; gene silencing Forward genetics: chemical mutagenesis Functional genomics and the central dogma Approaches to function; Functional genomics and DNA; …and RNA; …and protein Proteomic approaches to functional genomics CASP; protein-protein interactions; protein networks Perspective Albert Blakeslee (1874–1954) studied the effect of altered chromosome numbers on the phenotype of the jimson-weed Datura stramonium, a flowering plant. Introduction: Functional genomics Functional genomics is the genome-wide study of the function of DNA (including both genes and non-genic regions), as well as RNA and proteins encoded by DNA. The term “functional genomics” may apply to • the genome, transcriptome, or proteome • the use of high-throughput screens • the perturbation of gene function • the complex relationship of genotype and phenotype Functional genomics approaches to high throughput analyses Relationship between genotype and phenotype The genotype of an individual consists of the DNA that comprises the organism. The phenotype is the outward manifestation in terms of properties such as size, shape, movement, and physiology. We can consider the phenotype of a cell (e.g., a precursor cell may develop into a brain cell or liver cell) or the phenotype of an organism (e.g., a person may have a disease phenotype such as sickle‐cell anemia). -

Download on 20
bioRxiv preprint doi: https://doi.org/10.1101/850776; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Intramembrane protease RHBDL4 cleaves oligosaccharyltransferase subunits to target them for ER-associated degradation Julia D. Knopf1, Nina Landscheidt1, Cassandra L. Pegg2, Benjamin L. Schulz2, Nathalie Kühnle1, Chao-Wei Chao1, Simon Huck1 and Marius K. Lemberg1, # 1Centre for Molecular Biology of Heidelberg University (ZMBH), DKFZ-ZMBH Alliance, 69120 Heidelberg, Germany. 2School of Chemistry and Molecular Biosciences, ARC Training Centre for Biopharmaceutical Innovation, The University of Queensland, St Lucia QLD 4072, Australia. #Corresponding author: [email protected] Running title: RHBDL4 triggers ERAD of OST subunits Key words: Rhomboid serine protease, Rhbdd1, ubiquitin-dependent proteolysis, post- translational protein abundance control, N-linked glycosylation. Abbreviations ERAD, ER-associated degradation; OST, oligosacharyltransferase; TM, transmembrane; UIM, ubiquitin-interacting motif. Abstract The Endoplasmic Reticulum (ER)-resident intramembrane rhomboid protease RHBDL4 generates metastable protein fragments and together with the ER-associated degradation (ERAD) machinery provides a clearance mechanism for aberrant and surplus proteins. However, the endogenous substrate spectrum and with that the role of RHBDL4 in physiological ERAD is mainly unknown. Here, we use a substrate trapping approach in combination with quantitative proteomics to identify physiological RHBDL4 substrates. This revealed oligosacharyltransferase (OST) complex subunits such as the catalytic active subunit STT3A as substrates for the RHBDL4-dependent ERAD pathway. RHBDL4-catalyzed cleavage inactivates OST subunits by triggering dislocation into the cytoplasm and subsequent proteasomal degradation. -

Andrey L. Karamyshev
CURRICULUM VITAE Andrey L. Karamyshev Assistant Professor Department of Cell Biology and Biochemistry! Texas Tech University Health Sciences Center! 3601 4th Street, Mail Stop 6540! Lubbock, TX 79430 Phone: (806) 743-4102 Fax: (806) 743-2990 e-mail: [email protected] EDUCATION: Postdoctoral Studies: Department of Tumor Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan (Advisor: Dr. Yoshikazu Nakamura). Department of Medical Biochemistry and Genetics, College of Medicine, Texas A&M University, College Station, TX (Advisor: Dr. Arthur E. Johnson). Ph.D. in Biochemistry, Russian Academy of Sciences, Pushchino, Moscow Region, Russia. M.S., Biology, Kuban State University, Krasnodar, Russia. FIELDS OF SPECIALIZATION: Biochemistry, Molecular and Cellular Biology. RESEARCH INTERESTS Molecular mechanisms of human diseases. Post-transcriptional regulation of gene expression. Gene silencing and translational repression. mRNA and protein quality control. RNA stability and degradation. siRNAs/miRNAs. Prolactin, CFTR, secretory and membrane proteins. Protein translation, folding and transport. Protein-protein interactions. SCIENTIFIC PUBLICATIONS (see list below) 31 publications (total), including 25 full-length peer-reviewed papers in scientific journals, 5 articles in the Encyclopedia of Molecular Biology (Publisher: John Wiley & Sons, Inc., N.Y.), and chapter in a book. PRESENTATIONS (90 total, see lists below) Results were presented at various international and national meetings, conferences and symposia (58 presentations), as well as during invited or selected talks (32 talks) at different universities and meetings around the world. Andrey L. Karamyshev, Ph.D. RESEARCH EXPERIENCE AND POSITIONS 2016-present Assistant Professor (Tenure track), Department of Cell Biology and Biochemistry!, Texas Tech University Health Sciences Center!, Lubbock, TX. 2009-2016 Assistant Professor (Research track), Department of Physiology, UT Southwestern Medical Center at Dallas, Dallas, TX. -

Switch from Translation Initiation to Elongation Needs Not4 and Not5 Collaboration
bioRxiv preprint doi: https://doi.org/10.1101/850859; this version posted November 22, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Switch from translation initiation to elongation needs Not4 and Not5 collaboration George E Allen1°, Olesya O Panasenko1°, Zoltan Villanyi1,&, Marina Zagatti1, Benjamin Weiss1, 2 2 1* 5 Christine Polte , Zoya Ignatova , Martine A Collart 1Department of Microbiology and Molecular Medicine, Institute of Genetics and Genomics Geneva, Faculty of Medicine, University of Geneva, Switzerland 2Biochemistry and Molecular Biology, University of Hamburg, Germany °Equally contributing first authors 10 *Correspondence to: [email protected] &Current Address: Dept of Biochemistry and Molecular Biology, University of Szeged Abstract: Not4 and Not5 are crucial components of the Ccr4-Not complex with pivotal functions in mRNA metabolism. Both associate with ribosomes but mechanistic insights on their 15 function remain elusive. Here we determine that Not5 and Not4 synchronously impact translation initiation and Not5 alone alters translation elongation. Deletion of Not5 causes elongation defects in a codon-dependent fashion, increasing and decreasing the ribosome dwelling occupancy at minor and major codons, respectively. This larger difference in codons’ translation velocities alters translation globally and enables kinetically unfavorable processes 20 such as nascent chain deubiquitination to take place. In turn, this leads to abortive translation and favors protein aggregation. These findings highlight the global impact of Not4 and Not5 in controlling the speed of mRNA translation and transition from initiation to elongation. Summary: Not4 and Not5 regulate translation synchronously but distinguishably, facilitating 25 smooth transition from initiation to elongation Results and discussion The Ccr4-Not complex is a global regulator of mRNA metabolism in eukaryotic cells (for review see (1)). -
Download Validation Data
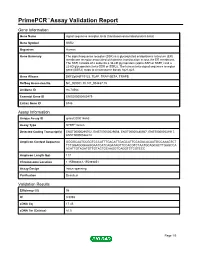
PrimePCR™Assay Validation Report Gene Information Gene Name signal sequence receptor, beta (translocon-associated protein beta) Gene Symbol SSR2 Organism Human Gene Summary The signal sequence receptor (SSR) is a glycosylated endoplasmic reticulum (ER) membrane receptor associated with protein translocation across the ER membrane. The SSR consists of 2 subunits a 34-kD glycoprotein (alpha-SSR or SSR1) and a 22-kD glycoprotein (beta-SSR or SSR2). The human beta-signal sequence receptor gene (SSR2) maps to chromosome bands 1q21-q23. Gene Aliases DKFZp686F19123, TLAP, TRAP-BETA, TRAPB RefSeq Accession No. NC_000001.10, NT_004487.19 UniGene ID Hs.74564 Ensembl Gene ID ENSG00000163479 Entrez Gene ID 6746 Assay Information Unique Assay ID qHsaCID0014663 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000295702, ENST00000529008, ENST00000480567, ENST00000531917, ENST00000526212 Amplicon Context Sequence GGGGCAATCCGGTCCCATTTGACATTGAGCATTCCAGACACAATGCCAAAGTCT TCTGGAGGGAAGGAATCATCAGATAGTTCCACGTCTAATGCAGCACTTGAGCCA ACATTGTAGATGTTGTACTGCAAGGTCAGGTCTCGTCCC Amplicon Length (bp) 117 Chromosome Location 1:155988061-155989851 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9998 cDNA Cq 17.45 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SSR2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

A Crosstalk Between the RNA Binding Protein Smaug and the Hedgehog Pathway Links Cell Signaling to Mrna Regulation in Drosophila Lucía Bruzzone
A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila Lucía Bruzzone To cite this version: Lucía Bruzzone. A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila. Cellular Biology. Université Sorbonne Paris Cité, 2018. English. NNT : 2018USPCC234. tel-02899776 HAL Id: tel-02899776 https://tel.archives-ouvertes.fr/tel-02899776 Submitted on 15 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparée à l’Université Paris Diderot Ecole doctorale HOB n° 561 Institut Jacques Monod / Equipe Développement, Signalisation et Trafic A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in Drosophila Lucía Bruzzone Thèse de doctorat de Biologie Dirigée par Anne Plessis Présentée et soutenue publiquement à Paris le 19 mars 2018 Président du jury: Alain Zider / Professeur Université Paris Diderot -

Glycan Engineering for Cell and Developmental Biology
View metadata, citation and similar papers at core.ac.uk brought to you by CORE HHS Public Access provided by Caltech Authors - Main Author manuscript Author ManuscriptAuthor Manuscript Author Cell Chem Manuscript Author Biol. Author Manuscript Author manuscript; available in PMC 2016 May 05. Published in final edited form as: Cell Chem Biol. 2016 January 21; 23(1): 108–121. doi:10.1016/j.chembiol.2015.12.007. Glycan Engineering for Cell and Developmental Biology Matthew E. Griffin1 and Linda C. Hsieh-Wilson1,* 1Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125, USA Abstract Cell-surface glycans are a diverse class of macromolecules that participate in many key biological processes, including cell-cell communication, development, and disease progression. Thus, the ability to modulate the structures of glycans on cell surfaces provides a powerful means not only to understand fundamental processes but also to direct activity and elicit desired cellular responses. Here, we describe methods to sculpt glycans on cell surfaces and highlight recent successes in which artificially engineered glycans have been employed to control biological outcomes such as the immune response and stem cell fate. Introduction Cell-surface glycans participate in many important processes throughout the lifespan of an organism, ranging from cell migration and tissue patterning to the immune response, disease progression, and cell death (Fuster and Esko, 2005; Häcker et al., 2005; Lichtenstein and Rabinovich, 2013; van Kooyk and Rabinovich, 2008). At a molecular level, glycans are often the first points of contact between cells, and they function by facilitating a variety of interactions both in cis (on the same cell) and in trans (on different cells). -

Gene Silencing: Double-Stranded RNA Mediated Mrna Degradation and Gene Inactivation
Cell Research (2001); 11(3):181-186 http://www.cell-research.com REVIEW Gene silencing: Double-stranded RNA mediated mRNA degradation and gene inactivation 1, 2 1 TANG WEI *, XIAO YAN LUO , VANESSA SANMUELS 1 North Carolina State University, Forest Biotechnology Group, Raleigh, NC 27695, USA 2 University of North Carolina, Department of Cell and Developmental Biology, Chapel Hill, NC 27599, USA ABSTRACT The recent development of gene transfer approaches in plants and animals has revealed that transgene can undergo silencing after integration in the genome. Host genes can also be silenced as a consequence of the presence of a homologous transgene. More and more investigations have demonstrated that double- stranded RNA can silence genes by triggering degradation of homologous RNA in the cytoplasm and by directing methylation of homologous nuclear DNA sequences. Analyses of Arabidopsis mutants and plant viral suppressors of silencing are unraveling RNA-silencing mechanisms and are assessing the role of me- thylation in transcriptional and posttranscriptional gene silencing. This review will focus on double-stranded RNA mediated mRNA degradation and gene inactivation in plants. Key words: Gene silencing, double-stranded RNA, methylation, homologous RNA, transgene. INTRODUCTION portant in consideration of its practical application The genome structure of plants can be altered by over the the past ten years[1-5]. Transgenes can genetic transformation. During the process of gene become silent after a long phase of expression, and transfer, Agrobacterium tumefaciens integrate part can sometimes silence the expression of homologous of their genome into the genome of susceptible elements located at ectopic positions in the genome. -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -

Newly Identified Gon4l/Udu-Interacting Proteins
www.nature.com/scientificreports OPEN Newly identifed Gon4l/ Udu‑interacting proteins implicate novel functions Su‑Mei Tsai1, Kuo‑Chang Chu1 & Yun‑Jin Jiang1,2,3,4,5* Mutations of the Gon4l/udu gene in diferent organisms give rise to diverse phenotypes. Although the efects of Gon4l/Udu in transcriptional regulation have been demonstrated, they cannot solely explain the observed characteristics among species. To further understand the function of Gon4l/Udu, we used yeast two‑hybrid (Y2H) screening to identify interacting proteins in zebrafsh and mouse systems, confrmed the interactions by co‑immunoprecipitation assay, and found four novel Gon4l‑interacting proteins: BRCA1 associated protein‑1 (Bap1), DNA methyltransferase 1 (Dnmt1), Tho complex 1 (Thoc1, also known as Tho1 or HPR1), and Cryptochrome circadian regulator 3a (Cry3a). Furthermore, all known Gon4l/Udu‑interacting proteins—as found in this study, in previous reports, and in online resources—were investigated by Phenotype Enrichment Analysis. The most enriched phenotypes identifed include increased embryonic tissue cell apoptosis, embryonic lethality, increased T cell derived lymphoma incidence, decreased cell proliferation, chromosome instability, and abnormal dopamine level, characteristics that largely resemble those observed in reported Gon4l/udu mutant animals. Similar to the expression pattern of udu, those of bap1, dnmt1, thoc1, and cry3a are also found in the brain region and other tissues. Thus, these fndings indicate novel mechanisms of Gon4l/ Udu in regulating CpG methylation, histone expression/modifcation, DNA repair/genomic stability, and RNA binding/processing/export. Gon4l is a nuclear protein conserved among species. Animal models from invertebrates to vertebrates have shown that the protein Gon4-like (Gon4l) is essential for regulating cell proliferation and diferentiation. -

Tannous A, Pisoni GB, Hebert DN, Molinari M. N-Linked Sugar
Seminars in Cell & Developmental Biology 41 (2015) 79–89 Contents lists available at ScienceDirect Seminars in Cell & Developmental Biology j ournal homepage: www.elsevier.com/locate/semcdb Review N-linked sugar-regulated protein folding and quality control in the ER a,1 b,1 a,∗ b,c,d,∗∗ Abla Tannous , Giorgia Brambilla Pisoni , Daniel N. Hebert , Maurizio Molinari a Department of Biochemistry and Molecular Biology, Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA 01003, USA b Università della Svizzera italiana, CH-6900 Lugano, Switzerland c Institute for Research in Biomedicine, Protein Folding and Quality Control, CH-6500 Bellinzona, Switzerland d Ecole Polytechnique Fédérale de Lausanne, School of Life Sciences, CH-1015 Lausanne, Switzerland a r t a b i s c l e i n f o t r a c t Article history: Asparagine-linked glycans (N-glycans) are displayed on the majority of proteins synthesized in the endo- Available online 19 December 2014 plasmic reticulum (ER). Removal of the outermost glucose residue recruits the lectin chaperone malectin possibly involved in a first triage of defective polypeptides. Removal of a second glucose promotes engage- Keywords: ment of folding and quality control machineries built around the ER lectin chaperones calnexin (CNX) Endoplasmic reticulum and calreticulin (CRT) and including oxidoreductases and peptidyl–prolyl isomerases. Deprivation of the N-glycosylation last glucose residue dictates the release of N-glycosylated polypeptides from the lectin chaperones. Cor- Protein folding and quality control rectly folded proteins are authorized to leave the ER. Non-native polypeptides are recognized by the ER Calnexin Calreticulin quality control key player UDP-glucose glycoprotein glucosyltransferase 1 (UGT1), re-glucosylated and re-addressed to the CNX/CRT chaperone binding cycle to provide additional opportunity for the protein to UDP-glucose glycoprotein glucosyltransferase 1 fold in the ER.