Disturbances Influence Trait Evolution in Pinus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Spatial Distribution and Historical Dynamics of Threatened Conifers of the Dalat Plateau, Vietnam
SPATIAL DISTRIBUTION AND HISTORICAL DYNAMICS OF THREATENED CONIFERS OF THE DALAT PLATEAU, VIETNAM A thesis Presented to The Faculty of the Graduate School At the University of Missouri In Partial Fulfillment Of the Requirements for the Degree Master of Arts By TRANG THI THU TRAN Dr. C. Mark Cowell, Thesis Supervisor MAY 2011 The undersigned, appointed by the dean of the Graduate School, have examined the thesis entitled SPATIAL DISTRIBUTION AND HISTORICAL DYNAMICS OF THREATENED CONIFERS OF THE DALAT PLATEAU, VIETNAM Presented by Trang Thi Thu Tran A candidate for the degree of Master of Arts of Geography And hereby certify that, in their opinion, it is worthy of acceptance. Professor C. Mark Cowell Professor Cuizhen (Susan) Wang Professor Mark Morgan ACKNOWLEDGEMENTS This research project would not have been possible without the support of many people. The author wishes to express gratitude to her supervisor, Prof. Dr. Mark Cowell who was abundantly helpful and offered invaluable assistance, support, and guidance. My heartfelt thanks also go to the members of supervisory committees, Assoc. Prof. Dr. Cuizhen (Susan) Wang and Prof. Mark Morgan without their knowledge and assistance this study would not have been successful. I also wish to thank the staff of the Vietnam Initiatives Group, particularly to Prof. Joseph Hobbs, Prof. Jerry Nelson, and Sang S. Kim for their encouragement and support through the duration of my studies. I also extend thanks to the Conservation Leadership Programme (aka BP Conservation Programme) and Rufford Small Grands for their financial support for the field work. Deepest gratitude is also due to Sub-Institute of Ecology Resources and Environmental Studies (SIERES) of the Institute of Tropical Biology (ITB) Vietnam, particularly to Prof. -

Encino En Guadalupe Y Calvo, Chihuahua Diversity and Vertical
Revista Mexicana de Ciencias Forestales Vol. 10 (53) May – June (2019) DOI: https://doi.org/10.29298/rmcf.v10i53.173 Article Diversidad y estructura vertical del bosque de pino– encino en Guadalupe y Calvo, Chihuahua Diversity and vertical structure of the pine-oak forest in Guadalupe y Calvo, Chihuahua Samuel Alberto García García1, Raúl Narváez Flores1, Jesús Miguel Olivas García1 y Javier Hernández Salas1 Resumen Se evaluaron áreas con y sin manejo forestal de la Umafor 0808 Guadalupe y Calvo, Chihuahua; gestionadas mediante el Método Mexicano de Ordenación de Bosques Irregulares (MMOBI). Se analizó y comparó la información de conglomerados del Inventario Nacional Forestal y de Suelos (2004-2009); 95 en masas con manejo y 27 sin manejo. Se determinó la estructura vertical por medio de la regeneración natural, pisos de altura de los árboles y posición sociológica. Las especies con distribución continua, desde el piso inferior de la regeneración hasta el piso arbóreo superior en el bosque con manejo fueron: Pinus durangensis, P. arizonica, P. ayacahuite, P. herrerae y P.engelmannii; mientras que, en el bosque sin manejo se registraron: P. durangensis y P. arizonica. Las principales diferencias entre los bosques estudiados correspondieron al promedio de altura en el piso arbóreo superior; en los bosques con manejo fue de 30.16 m y en los sin manejo, su valor fue de 21.86 m; además, se observó una mayor regeneración de P. durangensis en los primeros. Respecto a la diversidad de especies, no hubo diferencia significativa entre ambos tipos de bosque (P>0.05). Por lo anterior, se concluye que, de acuerdo con la información analizada, la regulación del aprovechamiento maderable con el MMOBI permite mantener la diversidad estructural y de especies, similar a la de un bosque natural sin manejo. -

Pines in the Arboretum
UNIVERSITY OF MINNESOTA MtJ ARBORETUM REVIEW No. 32-198 PETER C. MOE Pines in the Arboretum Pines are probably the best known of the conifers native to The genus Pinus is divided into hard and soft pines based on the northern hemisphere. They occur naturally from the up the hardness of wood, fundamental leaf anatomy, and other lands in the tropics to the limits of tree growth near the Arctic characteristics. The soft or white pines usually have needles in Circle and are widely grown throughout the world for timber clusters of five with one vascular bundle visible in cross sec and as ornamentals. In Minnesota we are limited by our cli tions. Most hard pines have needles in clusters of two or three mate to the more cold hardy species. This review will be with two vascular bundles visible in cross sections. For the limited to these hardy species, their cultivars, and a few hy discussion here, however, this natural division will be ignored brids that are being evaluated at the Arboretum. and an alphabetical listing of species will be used. Where neces Pines are readily distinguished from other common conifers sary for clarity, reference will be made to the proper groups by their needle-like leaves borne in clusters of two to five, of particular species. spirally arranged on the stem. Spruce (Picea) and fir (Abies), Of the more than 90 species of pine, the following 31 are or for example, bear single leaves spirally arranged. Larch (Larix) have been grown at the Arboretum. It should be noted that and true cedar (Cedrus) bear their leaves in a dense cluster of many of the following comments and recommendations are indefinite number, whereas juniper (Juniperus) and arborvitae based primarily on observations made at the University of (Thuja) and their related genera usually bear scalelikie or nee Minnesota Landscape Arboretum, and plant performance dlelike leaves that are opposite or borne in groups of three. -

Non-Native Trees and Large Shrubs for the Washington, D.C. Area
Green Spring Gardens 4603 Green Spring Rd ● Alexandria ● VA 22312 Phone: 703-642-5173 ● TTY: 703-803-3354 www.fairfaxcounty.gov/parks/greenspring NON - NATIVE TREES AND LARGE SHRUBS FOR THE WASHINGTON, D.C. AREA Non-native trees are some of the most beloved plants in the landscape due to their beauty. In addition, these trees are grown for the shade, screening, structure, and landscape benefits they provide. Deciduous trees, whose leaves die and fall off in the autumn, are valuable additions to landscapes because of their changing interest throughout the year. Evergreen trees are valued for their year-round beauty and shelter for wildlife. Evergreens are often grouped into two categories, broadleaf evergreens and conifers. Broadleaf evergreens have broad, flat leaves. They also may have showy flowers, such as Camellia oleifera (a large shrub), or colorful fruits, such as Nellie R. Stevens holly. Coniferous evergreens either have needle-like foliage, such as the lacebark pine, or scale-like foliage, such as the green giant arborvitae. Conifers do not have true flowers or fruits but bear cones. Though most conifers are evergreen, exceptions exist. Dawn redwood, for example, loses its needles each fall. The following are useful definitions: Cultivar (cv.) - a cultivated variety designated by single quotes, such as ‘Autumn Gold’. A variety (var.) or subspecies (subsp.), in contrast, is found in nature and is a subdivision of a species (a variety of Cedar of Lebanon is listed). Full Shade - the amount of light under a dense deciduous tree canopy or beneath evergreens. Full Sun - at least 6 hours of sun daily. -

Non-Wood Forest Products from Conifers
Page 1 of 8 NON -WOOD FOREST PRODUCTS 12 Non-Wood Forest Products From Conifers FAO - Food and Agriculture Organization of the United Nations The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. M-37 ISBN 92-5-104212-8 (c) FAO 1995 TABLE OF CONTENTS FOREWORD ACKNOWLEDGMENTS ABBREVIATIONS INTRODUCTION CHAPTER 1 - AN OVERVIEW OF THE CONIFERS WHAT ARE CONIFERS? DISTRIBUTION AND ABUNDANCE USES CHAPTER 2 - CONIFERS IN HUMAN CULTURE FOLKLORE AND MYTHOLOGY RELIGION POLITICAL SYMBOLS ART CHAPTER 3 - WHOLE TREES LANDSCAPE AND ORNAMENTAL TREES Page 2 of 8 Historical aspects Benefits Species Uses Foliage effect Specimen and character trees Shelter, screening and backcloth plantings Hedges CHRISTMAS TREES Historical aspects Species Abies spp Picea spp Pinus spp Pseudotsuga menziesii Other species Production and trade BONSAI Historical aspects Bonsai as an art form Bonsai cultivation Species Current status TOPIARY CONIFERS AS HOUSE PLANTS CHAPTER 4 - FOLIAGE EVERGREEN BOUGHS Uses Species Harvesting, management and trade PINE NEEDLES Mulch Decorative baskets OTHER USES OF CONIFER FOLIAGE CHAPTER 5 - BARK AND ROOTS TRADITIONAL USES Inner bark as food Medicinal uses Natural dyes Other uses TAXOL Description and uses Harvesting methods Alternative -

Current and Potential Spatial Distribution of Six Endangered Pine Species of Mexico: Towards a Conservation Strategy
Article Current and Potential Spatial Distribution of Six Endangered Pine Species of Mexico: Towards a Conservation Strategy Martin Enrique Romero-Sanchez * , Ramiro Perez-Miranda, Antonio Gonzalez-Hernandez, Mario Valerio Velasco-Garcia , Efraín Velasco-Bautista and Andrés Flores National Institute on Forestry, Agriculture and Livestock Research, Progreso 5, Barrio de Santa Catarina, Coyoacan, 04010 Mexico City, Mexico; [email protected] (R.P.-M.); [email protected] (A.G.-H.); [email protected] (M.V.V.-G.); [email protected] (E.V.-B.); fl[email protected] (A.F.) * Correspondence: [email protected]; Tel.: +52-553-626-8698 Received: 24 October 2018; Accepted: 6 December 2018; Published: 12 December 2018 Abstract: Mexico is home to the highest species diversity of pines: 46 species out of 113 reported around the world. Within the great diversity of pines in Mexico, Pinus culminicola Andresen et Beaman, P. jaliscana Perez de la Rosa, P. maximartinenzii Rzed., P. nelsonii Shaw, P. pinceana Gordon, and P. rzedowskii Madrigal et M. Caball. are six catalogued as threatened or endangered due to their restricted distribution and low population density. Therefore, they are of special interest for forest conservation purposes. In this paper, we aim to provide up-to-date information on the spatial distribution of these six pine species according to different historical registers coming from different herbaria distributed around the country by using spatial modeling. Therefore, we recovered historical observations of the natural distribution of each species and modelled suitable areas of distribution according to environmental requirements. Finally, we evaluated the distributions by contrasting changes of vegetation in the period 1991–2016. -

Phytosociological Analysis of Pine Forest at Indus Kohistan, Kpk, Pakistan
Pak. J. Bot., 48(2): 575-580, 2016. PHYTOSOCIOLOGICAL ANALYSIS OF PINE FOREST AT INDUS KOHISTAN, KPK, PAKISTAN ADAM KHAN1, MOINUDDIN AHMED2, MUHAMMAD FAHEEM SIDDIQUI*3, JAVED IQBAL1 AND MUHAMMAD WAHAB4 1Laboratory of plant ecology and Dendrochronology, Department of Botany, Federal Urdu University, Gulshan-e-Iqbal Campus Karachi, Pakistan 2Department of Earth and Environmental Systems, 600 Chestnut Street Indiana State University, Terre Haute, IN, USA 3Department of Botany, University of Karachi, Karachi-75270, Pakistan 4Institute of Botany, Chinese Academy of Sciences, Beijing, China *Corresponding author’s email: [email protected] Abstract The study was carried out to describe the pine communities at Indus Kohistan valley in quantitative term. Thirty stands of relatively undisturbed vegetation were selected for sampling. Quantitative sampling was carried out by Point Centered Quarter (PCQ) method. Seven tree species were common in the Indus Kohistan valley. Cedrus deodara was recorded from twenty eight different locations and exhibited the highest mean importance value while Pinus wallichiana was recorded from 23 different locations and exhibited second highest mean importance value. Third most occurring species was Abies pindrow that attained the third highest mean importance value and Picea smithiana was recorded from eight different locations and attained fourth highest importance value while it was first dominant in one stand and second dominant in four stands. Pinus gerardiana, Quercus baloot and Taxus fuana were the rare species in this area, these species attained low mean importance value. Six communities and four monospecific stands of Cedrus deodara were recognized. Cedrus-Pinus community was the most occurring community, which was recorded from 13 different stands. -
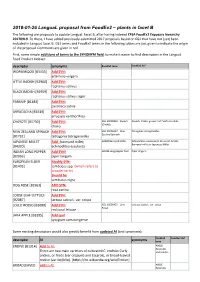
2018-01-26 Langual Proposal from Foodex2 – Plants in Facet B
2018-01-26 LanguaL proposal from FoodEx2 – plants in facet B The following are proposals to update LanguaL Facet B, after having indexed EFSA FoodEx2 Exposure hierarchy 20170919. To these, I have added previously-submitted 2017 proposals based on GS1 that have not (yet) been included in LanguaL facet B. GS1 terms and FoodEx2 terms in the following tables are just given to indicate the origin of the proposal. Comments are given in red. First, some simple additions of terms to the SYNONYM field, to make it easier to find descriptors in the LanguaL Food Product Indexer: descriptor synonyms FoodEx2 term FoodEx2 def WORMWOOD [B3433] Add SYN: artemisia vulgaris LITTLE RADISH [B2960] Add SYN: raphanus sativus BLACK RADISH [B2959] Add SYN: raphanus sativus niger PARSNIP [B1483] Add SYN: pastinaca sativa ARRACACHA [B3439] Add SYN: arracacia xanthorrhiza CHAYOTE [B1730] Add SYN: GS1 10006356 - Squash Squash, Choko, grown from Sechium edule (Choko) choko NEW ZEALAND SPINACH Add SYN: GS1 10006427 - New- Tetragonia tetragonoides Zealand Spinach [B1732] tetragonia tetragonoides JAPANESE MILLET Add : barnyard millet; A000Z Barnyard millet Echinochloa esculenta (A. Braun) H. Scholz, Barnyard millet or Japanese Millet. [B4320] echinochloa esculenta INDIAN LONG PEPPER Add SYN! A019B Long pepper fruit Piper longum [B2956] piper longum EUROPEAN ELDER Modify SYN: [B1403] sambucus spp. (which refers to broader term) Should be sambucus nigra DOG ROSE [B2961] ADD SYN: rosa canina LOOSE LEAF LETTUCE Add SYN: [B2087] lactusa sativa L. var. crispa LOLLO ROSSO [B2088] Add SYN: GS1 10006425 - Lollo Lactuca sativa L. var. crispa Rosso red coral lettuce JAVA APPLE [B3395] Add syn! syzygium samarangense Some existing descriptors would also greatly benefit from updated AI (and synonyms): FoodEx2 FoodEx2 def descriptor AI synonyms term ENDIVE [B1314] Add to AI: A00LD Escaroles There are two main varieties of cultivated C. -

Biodiversity Conservation in Botanical Gardens
AgroSMART 2019 International scientific and practical conference ``AgroSMART - Smart solutions for agriculture'' Volume 2019 Conference Paper Biodiversity Conservation in Botanical Gardens: The Collection of Pinaceae Representatives in the Greenhouses of Peter the Great Botanical Garden (BIN RAN) E M Arnautova and M A Yaroslavceva Department of Botanical garden, BIN RAN, Saint-Petersburg, Russia Abstract The work researches the role of botanical gardens in biodiversity conservation. It cites the total number of rare and endangered plants in the greenhouse collection of Peter the Great Botanical garden (BIN RAN). The greenhouse collection of Pinaceae representatives has been analysed, provided with a short description of family, genus and certain species, presented in the collection. The article highlights the importance of Pinaceae for various industries, decorative value of plants of this group, the worth of the pinaceous as having environment-improving properties. In Corresponding Author: the greenhouses there are 37 species of Pinaceae, of 7 geni, all species have a E M Arnautova conservation status: CR -- 2 species, EN -- 3 species, VU- 3 species, NT -- 4 species, LC [email protected] -- 25 species. For most species it is indicated what causes depletion. Most often it is Received: 25 October 2019 the destruction of natural habitats, uncontrolled clearance, insect invasion and diseases. Accepted: 15 November 2019 Published: 25 November 2019 Keywords: biodiversity, botanical gardens, collections of tropical and subtropical plants, Pinaceae plants, conservation status Publishing services provided by Knowledge E E M Arnautova and M A Yaroslavceva. This article is distributed under the terms of the Creative Commons 1. Introduction Attribution License, which permits unrestricted use and Nowadays research of biodiversity is believed to be one of the overarching goals for redistribution provided that the original author and source are the modern world. -

Hybridization and Evolution in the Genus Pinus
Hybridization and Evolution in the Genus Pinus Baosheng Wang Department of Ecology and Environmental Science Umeå 2013 Hybridization and Evolution in the Genus Pinus Baosheng Wang Department of Ecology and Environmental Science Umeå University, Umeå, Sweden 2013 This work is protected by the Swedish Copyright Legislation (Act 1960:729) Copyright©Baosheng Wang ISBN: 978-91-7459-702-8 Cover photo: Jian-Feng Mao Printed by: Print&Media Umeå, Sweden 2013 List of Papers This thesis is a summary and discussion of the following papers, which are referred to by their Roman numerals. I. Wang, B. and Wang, X.R. Mitochondrial DNA capture and divergence in Pinus provide new insights into the evolution of the genus. Submitted Manuscript II. Wang, B., Mao, J.F., Gao, J., Zhao, W. and Wang, X.R. 2011. Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata. Molecular Ecology 20: 3796-3811. III. Gao, J., Wang, B., Mao, J.F., Ingvarsson, P., Zeng, Q.Y. and Wang, X.R. 2012. Demography and speciation history of the homoploid hybrid pine Pinus densata on the Tibetan Plateau. Molecular Ecology 21: 4811–4827. IV. Wang, B., Mao, J.F., Zhao, W. and Wang, X.R. 2013. Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnannensis. PLoS One. 8: e67345. doi:10.1371/journal.pone.0067345 V. Wang, B., Mahani, M.K., Ng, W.L., Kusumi, J., Phi, H.H., Inomata, N., Wang, X.R. and Szmidt, A.E. Extremely low nucleotide polymorphism in Pinus krempfii Lecomte, a unique flat needle pine endemic to Vietnam. -

Keel, S. 2005. Caribbean Ecoregional Assessment Cuba Terrestrial
CARIBBEAN ECOREGIONAL ASSESSMENT Cuba Terrestrial Report July 8, 2005 Shirley Keel INTRODUCTION Physical Features Cuba is the largest country in the Caribbean, with a total area of 110,922 km2. The Cuba archipelago consists of the main island (105,007 km2), Isla de Pinos (2,200 km2), and more than one thousand cays (3,715 km2). Cuba’s main island, oriented in a NW-SE direction, has a varied orography. In the NW the major mountain range is the Guaniguanico Massif stretching from west to east with two mountain chains of distinct geological ages and composition—Sierra de los Organos of ancient Jurassic limestone deposited on slaty sandstone, and Sierra del Rosario, younger and highly varied in geological structure. Towards the east lie the low Hills of Habana- Matanzas and the Hills of Bejucal-Madruga-Limonar. In the central part along the east coast are several low hills—from north to south the Mogotes of Caguaguas, Loma Cunagua, the ancient karstic range of Sierra de Cubitas, and the Maniabón Group; while along the west coast rises the Guamuhaya Massif (Sierra de Escambray range) and low lying Sierra de Najasa. In the SE, Sierra Maestra and the Sagua-Baracoa Massif form continuous mountain ranges. The high ranges of Sierra Maestra stretch from west to east with the island’s highest peak, Pico Real (Turquino Group), reaching 1,974 m. The complex mountain system of Sagua-Baracoa consists of several serpentine mountains in the north and plateau-like limestone mountains in the south. Low limestone hills, Sierra de Casas and Sierra de Caballos are situated in the northeastern part of Isla de Pinos (Borhidi, 1991). -

Songbird Ecology in Southwestern Ponderosa
This file was created by scanning the printed publication. Errors identified by the software have been corrected; however, some errors may remain. Chapter 1 Ecology of Southwestern Ponderosa Pine Forests William H. Moir, Brian Geils, Mary Ann Benoit, and Dan Scurlock describes natural and human induced changes in the com- What Is Ponderosa Pine Forest position and structure of these forests. and Why Is It Important? Forests dominated by ponderosa pine (Pinus ponderosa Paleoecology var. scopulorurn) are a major forest type of western North America (figure 1; Steele 1988; Daubenmire 1978; Oliver and Ryker 1990). In this publication, a ponderosa pine The oldest remains of ponderosa pine in the Western forest has an overstory, regardless of successional stage, United States are 600,000 year old fossils found in west dominated by ponderosa pine. This definition corresponds central Nevada. Examination of pack rat middens in New to the interior ponderosa pine cover type of the Society of Mexico and Texas, shows that ponderosa pine was absent American Foresters (Eyre1980). At lower elevations in the during the Wisconsin period (about 10,400 to 43,000 years mountainous West, ponderosa pine forests are generally ago), although pinyon-juniper woodlands and mixed co- bordered by grasslands, pinyon-juniper woodlands, or nifer forests were extensive (Betancourt 1990). From the chaparral (shrublands). The ecotone may be wide or nar- late Pleistocene epoch (24,000 years ago) to the end of the row, and a ponderosa pine forest is recognized when the last ice age (about 10,400 years ago), the vegetation of the overstory contains at least 5 percent ponderosa pine (USFS Colorado Plateau moved southward or northward with 1986).