Molecular Characterisation of Two Ornithodoros Savignyi Enzyme Isoforms from the 5
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome Is Evolutionarily Conserved
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.28.317750; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome is Evolutionarily Conserved Authors Peter Kubiniok†1, Ana Marcu†2,3, Leon Bichmann†2,4, Leon Kuchenbecker4, Heiko Schuster1,5, David Hamelin1, Jérome Despault1, Kevin Kovalchik1, Laura Wessling1, Oliver Kohlbacher4,7,8,9,10 Stefan Stevanovic2,3,6, Hans-Georg Rammensee2,3,6, Marian C. Neidert11, Isabelle Sirois1, Etienne Caron1,12* Affiliations *Corresponding and Leading author: Etienne Caron ([email protected]) †Equal contribution to this work 1CHU Sainte-Justine Research Center, Montreal, QC H3T 1C5, Canada 2Department of Immunology, Interfaculty Institute for Cell Biology, University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 3Cluster of Excellence iFIT (EXC 2180) "Image-Guided and Functionally Instructed Tumor Therapies", University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 4Applied Bioinformatics, Dept. of Computer Science, University of Tübingen, Tübingen, Baden- Württemberg, 72074, Germany. 5Immatics Biotechnologies GmbH, Tübingen, 72076, Baden-Württemberg, Germany. 6DKFZ Partner Site Tübingen, German Cancer Consortium (DKTK), Tübingen, Baden- Württemberg, 72076, Germany. 7Institute for Bioinformatics and Medical Informatics, -

Babela Massiliensis, a Representative of a Widespread Bacterial
Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, Samia Benamar, Didier Raoult, Eugene V. Koonin, Bernard La Scola To cite this version: Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, et al.. Babela mas- siliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biology Direct, BioMed Central, 2015, 10 (13), 10.1186/s13062-015-0043-z. hal-01217089 HAL Id: hal-01217089 https://hal-amu.archives-ouvertes.fr/hal-01217089 Submitted on 19 Oct 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Pagnier et al. Biology Direct (2015) 10:13 DOI 10.1186/s13062-015-0043-z RESEARCH Open Access Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier1, Natalya Yutin2, Olivier Croce1, Kira S Makarova2, Yuri I Wolf2, Samia Benamar1, Didier Raoult1, Eugene V Koonin2 and Bernard La Scola1* Abstract Background: Only a small fraction of bacteria and archaea that are identifiable by metagenomics can be grown on standard media. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Novel Missense Mutation in PTPN22 in a Chinese Pedigree With
Gong et al. BMC Endocrine Disorders (2018) 18:76 https://doi.org/10.1186/s12902-018-0305-8 RESEARCHARTICLE Open Access Novel missense mutation in PTPN22 in a Chinese pedigree with Hashimoto’s thyroiditis Licheng Gong1†, Beihong Liu2,3†, Jing Wang4, Hong Pan3, Anhui Qi2,3, Siyang Zhang2,3, Jinyi Wu1, Ping Yang1* and Binbin Wang3,4,5* Abstract Background: Hashimoto’s thyroiditis is a complex autoimmune thyroid disease, the onset of which is associated with environmental exposures and specific susceptibility genes. Its incidence in females is higher than its incidence in males. Thus far, although some susceptibility loci have been elaborated, including PTPN22, FOXP3, and CD25, the aetiology and pathogenesis of Hashimoto’s thyroiditis remains unclear. Methods: Four affected members from a Chinese family with Hashimoto’s thyroiditis were selected for whole-exome sequencing. Missense, nonsense, frameshift, or splicing-site variants shared by all affected members were identified after frequency filtering against public and internal exome databases. Segregation analysis was performed by Sanger sequencing among all members with available DNA. Results: We identified a missense mutation in PTPN22 (NM_015967.5; c. 77A > G; p.Asn26Ser) using whole-exome sequencing. PTPN22 is a known susceptibility gene associated with increased risks of multiple autoimmune diseases. Cosegregation analysis confirmed that all patients in this family, all of whom were female, carried the mutation. All public and private databases showed that the missense mutation was extremely rare. Conclusions: We found a missense mutation in PTPN22 in a Chinese HT pedigree using whole-exome sequencing. Our study, for the first time, linked a rare variant of PTPN22 to Hashimoto’s thyroiditis, providing further evidence of the disease-causing or susceptibility role of PTPN22 in autoimmune thyroid disease. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Diplomarbeit
DIPLOMARBEIT Titel der Diplomarbeit „The influence of free fatty acids on the development of liver inflammation“ Verfasser Mario Kuttke, B.Sc. angestrebter akademischer Grad Magister der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. Studienblatt: A 490 Studienrichtung lt. Studienblatt: Diplomstudium Molekulare Biologie Betreuerin / Betreuer: A.o.Univ.-Prof.Dipl.-Ing.Dr. Marcela Hermann Danksagung Zuerst möchte ich mich bei a.o.Univ.-Prof. Dipl.-Ing. Dr. Marcela Hermann für die Betreuung meiner Diplomarbeit bedanken. Besonderer Dank gilt a.o.Univ-Prof. Dr. Bettina Grasl-Kraupp für die fachliche Unterstützung und Betreuung während der praktischen Durchführung der Arbeit. Weiters bedanke ich mich bei Sandra Sagmeister, Therese Böhm, Nora Bintner, Waltraud Schrottmaier, Melanie Pichlbauer, Marzieh Nejabat, Teresa Riegler, Bettina Wingelhofer und Christiane Maier für die ausgezeichnete Zusammenarbeit im Labor und die Unterstützung in allen Lebenslagen. Birgit Mir-Karner, Helga Koudelka und Krystyna Bukowska danke ich für ihre Hilfsbereitschaft und für die kollegiale Zusammenarbeit. Mein größter Dank gilt meinen Eltern, Ursula und Heinz, und meiner Großmutter, Theresia, die mir mein Studium ermöglicht und mich immer unterstützt haben, sowie meinem Bruder, Alex, der in allen Lebenslagen für mich da ist. Table of Contents TABLE OF CONTENTS INTRODUCTION ............................................................................................................................................. 4 HEPATOCELLULAR CARCINOMA (HCC) -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -

Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts
UC Irvine UC Irvine Previously Published Works Title Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts. Permalink https://escholarship.org/uc/item/5534264q Authors Li, Ye Salazar, Joelle K He, Yingshu et al. Publication Date 2020 DOI 10.3389/fmicb.2020.582202 Peer reviewed eScholarship.org Powered by the California Digital Library University of California fmicb-11-582202 October 19, 2020 Time: 10:46 # 1 ORIGINAL RESEARCH published: 23 October 2020 doi: 10.3389/fmicb.2020.582202 Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts Ye Li1, Joelle K. Salazar2, Yingshu He1, Prerak Desai3, Steffen Porwollik3, Weiping Chu3, Palma-Salgado Sindy Paola4, Mary Lou Tortorello2, Oscar Juarez5, Hao Feng4, Michael McClelland3* and Wei Zhang1* 1 Department of Food Science and Nutrition, Illinois Institute of Technology, Bedford Park, IL, United States, 2 Division of Food Processing Science and Technology, U.S. Food and Drug Administration, Bedford Park, IL, United States, 3 Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States, 4 Department of Food Science and Human Nutrition, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5 Department Edited by: of Biology, Illinois Institute of Technology, Chicago, IL, United States Chrysoula C. Tassou, Institute of Technology of Agricultural Products, Hellenic Agricultural Salmonella enterica subspecies I (ssp 1) is the leading cause of hospitalizations and Organization, Greece deaths due to known bacterial foodborne pathogens in the United States and is Reviewed by: frequently implicated in foodborne disease outbreaks associated with spices and nuts. -

Towards the Elucidation of Orphan Lysosomal Transporters Quentin Verdon
Towards the Elucidation of Orphan Lysosomal Transporters Quentin Verdon To cite this version: Quentin Verdon. Towards the Elucidation of Orphan Lysosomal Transporters. Cancer. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS144. tel-01827233 HAL Id: tel-01827233 https://tel.archives-ouvertes.fr/tel-01827233 Submitted on 2 Jul 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS144 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY PREPAREE A L’UNIVERSITE PARIS-SUD ECOLE DOCTORALE N°568 BIOSIGNE | Signalisations et réseaux intégratifs en biologie Spécialité de doctorat : aspects moléculaires et cellulaires de la biologie Par Mr Quentin Verdon Towards the elucidation of orphan lysosomal transporters: several shots on target and one goal Thèse présentée et soutenue à Paris le 29/06/2016 » : Composition du Jury : Mr Le Maire Marc Professeur, Université Paris-Sud Président Mr Birman Serge Directeur de recherche, CNRS Rapporteur Mr Murray James Assistant professor, Trinity college Dublin Rapporteur Mr Goud Bruno Directeur de recherche, CNRS Examinateur Mr Gasnier Bruno Directeur de recherche, CNRS Directeur de thèse Mme Sagné Corinne Chargée de recherche, INSERM Co-directeur de thèse Table of contents Remerciements (acknowledgements) 6 Abbreviations 7 Abstracts 10 Introduction 12 1 Physiology of lysosomes 12 1.1 Discovery and generalities 12 1.2 Degradative function 13 1.3. -

Interplay Between Uridylation and Deadenylation During Mrna Degradation in Arabidopsis Thaliana
$FNQRZOHGJPHQWV &ŝƌƐƚ͕/ǁŽƵůĚůŝŬĞƚŽƚŚĂŶŬaƚĢƉĄŶŬĂsĂŸĄēŽǀĄ ͕ŶĚƌĞĂƐtĂĐŚƚĞƌĂŶĚ&ĂďŝĞŶŶĞDĂƵdžŝŽŶĨŽƌŚĂǀŝŶŐ ĂĐĐĞƉƚĞĚƚŽĞǀĂůƵĂƚĞŵLJǁŽƌŬ͘DĞƌĐŝĠŐĂůĞŵĞŶƚ ĂƵ>ĂďdžEĞƚZEĚ͛ĂǀŽŝƌĨŝŶĂŶĐĠŵĂƚŚğƐĞ͘ :͛ĂŝŵĞƌĂŝƐƌĞŵĞƌĐŝĞƌŵŽŶĚŝƌĞĐƚĞƵƌĚĞƚŚğƐĞ ͕ŽŵŝŶŝƋƵĞ'ĂŐůŝĂƌĚŝ;ͨ'ĂŐͩͿ͕ ĚĞŵ͛ĂǀŽŝƌĂĐĐƵĞŝůůŝ ĂƵƐĞŝŶĚĞƐŽŶĠƋƵŝƉĞĞƚĚĞŵ͛ĂǀŽŝƌƐŽƵƚĞŶƵ ƚŽƵƚĂƵůŽŶŐĚĞŵĂƚŚğƐĞ͘:ĞƚĞƌĞŵĞƌĐŝĞƉŽƵƌƚŽŶ ĞdžƉĞƌƚŝƐĞ͕ƚ ĞƐƉƌĠĐŝĞƵdžĐŽŶƐĞŝůƐĞƚůĞƐ;ůŽŶŐƵĞƐ͙ ͿĚŝƐĐƵƐƐŝŽŶƐƐĐŝĞŶƚŝĨŝƋƵĞƐƋƵŝŵ͛ŽŶƚďĞĂƵĐŽƵƉ ĂƉƉŽƌƚĠĞƚĂŝĚĠăĚĠǀĞůŽƉƉĞƌŵŽŶĞƐƉƌŝƚĐƌŝƚŝƋƵĞĞƚƐĐŝĞŶƚŝĨŝƋƵĞ͘ hŶŐƌĂŶĚŵĞƌĐŝă,ĠůğŶĞƵďĞƌƉŽƵƌƐĂŐĞŶƚŝůůĞƐƐĞĞƚƐĂƉĂƚŝĞŶĐĞĚƵƌĂŶƚĐĞƐϰĂŶŶĠĞƐ͘DĞƌĐŝƉŽƵƌ ƚŽŶ ƐŽƵƚŝĞŶ ŵŽƌĂů Ğƚ ŝŶƚĞůůĞĐƚƵĞů͕ ƚƵ ĂƐ ƚŽƵũŽƵƌƐ ĠƚĠ ůă ƉŽƵƌ ƌĠƉŽŶĚƌĞ ă ŵĞƐ ŝŶŶŽŵďƌĂďůĞƐ ƋƵĞƐƚŝŽŶƐĞƚƚƵĂƐƚŽƵũŽƵƌƐƐƵŵ͛ĞŶĐŽƵƌĂŐĞƌƋƵĂŶĚĕĂŶ͛ĂůůĂŝƚƉĂƐƚƌŽƉ͘ ƚŵĞƌĐŝĂƵƐƐŝƉŽƵƌƚĂ ƉĂƚŝĞŶĐĞ Ğƚ ƚ ĞƐ ďŽŶƐ ĐŽŶƐĞŝůƐ ůŽƌƐ ĚĞ ů͛ĠĐƌŝƚƵƌĞ ĚĞ ĐĞƚƚĞ ƚŚğƐĞ͘ dƵ ĞƐ ƵŶĞ ƉĞƌƐŽŶŶĞ Ğƚ ƵŶĞ ƐĐŝĞŶƚŝĨŝƋƵĞ ƌĞŵĂƌƋƵĂďůĞ͕ ũĞ ƐƵŝƐ ĐŽŶĨŝĂŶƚĞ ƋƵĞ ƚƵ ŝƌĂƐ ůŽŝŶ ĚĂŶƐ ƚĂ ǀŝĞ ƉĞƌƐŽŶŶĞůůĞ Ğƚ ƉƌŽĨĞƐƐŝŽŶŶĞůůĞ͊ ŝŶ ďĞƐŽŶĚĞƌĞƌ ĂŶŬ Őŝůƚ ,ĞŝŬĞ Ĩƺƌ ŝŚƌĞ ĞƚƌĞƵƵŶŐ ƵŶĚ ŝŚƌĞ ŐƵƚĞŶ ZĂƚƐĐŚůćŐĞ͘ /ĐŚ ĚĂŶŬĞ Ěŝƌ ĂƵĨƌŝĐŚƚŝŐĨƺƌĚĞŝŶĞŶŬŽŶƐƚƌƵŬƚŝǀĞŶĞŝƚƌĂŐƵŶĚĚĞŝŶĞƵŶĞƌŵƺĚůŝĐŚĞhŶƚĞƌƐƚƺƚnjƵŶŐ͕ĚŝĞŵŝƌďĞŝĚĞƌ hŵƐĞƚnjƵŶŐĚŝĞƐĞƐDĂŶƵƐŬƌŝƉƚƐƐĞŚƌŐĞŚŽůĨĞŶŚĂďĞŶ͘,ćƚƚĞƐƚĚƵŵŝĐŚĚĂŵĂůƐŶŝĐŚƚĂůƐWƌĂŬƚŝŬĂŶƚŝŶ ŐĞŶŽŵŵĞŶ͕ǁćƌĞŝĐŚǁŽŚůũĞƚnjƚŶŝĐŚƚŚŝĞƌ͘ƵďŝƐƚĞŝŶĞƐĞŚƌĂƵĨŵĞƌŬƐĂŵĞƵŶĚŚŝůĨƐďĞƌĞŝƚĞWĞƌƐŽŶ͕ ĚŝĞŝĐŚƐĞŚƌƐĐŚćƚnjĞƵŶĚŶŝĐŚƚƐŽƐĐŚŶĞůůǀĞƌŐĞƐƐĞŶǁĞƌĚĞ͘ DĞƌĐŝăDĂƌůğŶĞĞƚ,ĠůğŶĞĚ͛ĂǀŽŝƌĠƚĠ ůă͊sŽƵƐġƚĞƐĚĞƐĂŵŝĞƐƉƌĠĐŝĞƵƐĞƐƋƵĞũĞŶĞƐƵŝƐƉĂƐƉƌġƚĞ ăŽƵďůŝĞƌ͊DĞƌĐŝDĂƌůğŶĞƉŽƵƌƚŽŶŐƌĂŝŶĚĞĨŽůŝĞ͕ƚĂďŽŶŶĞŚƵŵĞƵƌĞƚƚŽŶŚŽŶŶġƚĞƚĠ͕ĞƚƉŽƵƌƚŽƵƐ ĐĞƐ ŵŽŵĞŶƚƐ ƉĂƐƐĠƐ ĂƵ ůĂďŽ Ğƚ ƐƵƌƚŽƵƚ ĞŶ ĚĞŚŽƌƐ ĚƵ ůĂďŽ ĂǀĞĐ ,ĠůğŶĞ ͊ DĞƌĐŝ ,ĠůğŶĞ͕ ũĞ -

Exoribonuclease Nibbler Shapes the 3″ Ends of Micrornas
Current Biology 21, 1878–1887, November 22, 2011 ª2011 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2011.09.034 Article The 30-to-50 Exoribonuclease Nibbler Shapes the 30 Ends of MicroRNAs Bound to Drosophila Argonaute1 Bo W. Han,1 Jui-Hung Hung,2 Zhiping Weng,2 precursor miRNAs (pre-miRNAs) [8]. Pre-miRNAs comprise Phillip D. Zamore,1,* and Stefan L. Ameres1,* a single-stranded loop and a partially base-paired stem whose 1Howard Hughes Medical Institute and Department of termini bear the hallmarks of RNase III processing: a two-nucle- Biochemistry and Molecular Pharmacology otide 30 overhang, a 50 phosphate, and a 30 hydroxyl group. 2Program in Bioinformatics and Integrative Biology Nuclear pre-miRNAs are exported by Exportin 5 to the cyto- University of Massachusetts Medical School, plasm, where the RNase III enzyme Dicer liberates w22 nt 364 Plantation Street, Worcester, MA 01605, USA mature miRNA/miRNA* duplexes from the pre-miRNA stem [9–12]. Like all Dicer products, miRNA duplexes contain two- nucleotide 30 overhangs, 50 phosphate, and 30 hydroxyl groups. Summary In flies, Dicer-1 cleaves pre-miRNAs to miRNAs, whereas Dicer-2 converts long double-stranded RNA (dsRNA) into Background: MicroRNAs (miRNAs) are w22 nucleotide (nt) small interfering RNAs (siRNAs), which direct RNA interference small RNAs that control development, physiology, and pathol- (RNAi), a distinct small RNA silencing pathway required for ogy in animals and plants. Production of miRNAs involves the host defense against viral infection and somatic transposon sequential processing of primary hairpin-containing RNA poly- mobilization, as well as gene silencing triggered by exogenous merase II transcripts by the RNase III enzymes Drosha in the dsRNA [13, 14]. -
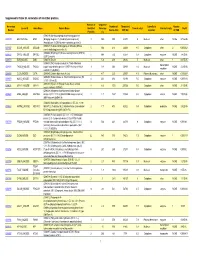
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase