Characterization of the Mammalian RNA Exonuclease 5/NEF-Sp As a Testis-Specific Nuclear 3′′′′′ → 5′′′′′ Exoribonuclease
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts
UC Irvine UC Irvine Previously Published Works Title Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts. Permalink https://escholarship.org/uc/item/5534264q Authors Li, Ye Salazar, Joelle K He, Yingshu et al. Publication Date 2020 DOI 10.3389/fmicb.2020.582202 Peer reviewed eScholarship.org Powered by the California Digital Library University of California fmicb-11-582202 October 19, 2020 Time: 10:46 # 1 ORIGINAL RESEARCH published: 23 October 2020 doi: 10.3389/fmicb.2020.582202 Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts Ye Li1, Joelle K. Salazar2, Yingshu He1, Prerak Desai3, Steffen Porwollik3, Weiping Chu3, Palma-Salgado Sindy Paola4, Mary Lou Tortorello2, Oscar Juarez5, Hao Feng4, Michael McClelland3* and Wei Zhang1* 1 Department of Food Science and Nutrition, Illinois Institute of Technology, Bedford Park, IL, United States, 2 Division of Food Processing Science and Technology, U.S. Food and Drug Administration, Bedford Park, IL, United States, 3 Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States, 4 Department of Food Science and Human Nutrition, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5 Department Edited by: of Biology, Illinois Institute of Technology, Chicago, IL, United States Chrysoula C. Tassou, Institute of Technology of Agricultural Products, Hellenic Agricultural Salmonella enterica subspecies I (ssp 1) is the leading cause of hospitalizations and Organization, Greece deaths due to known bacterial foodborne pathogens in the United States and is Reviewed by: frequently implicated in foodborne disease outbreaks associated with spices and nuts. -
Enzymes and Rna Complexes
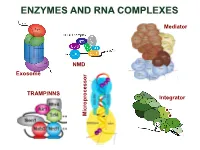
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

Exoribonuclease Nibbler Shapes the 3″ Ends of Micrornas
Current Biology 21, 1878–1887, November 22, 2011 ª2011 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2011.09.034 Article The 30-to-50 Exoribonuclease Nibbler Shapes the 30 Ends of MicroRNAs Bound to Drosophila Argonaute1 Bo W. Han,1 Jui-Hung Hung,2 Zhiping Weng,2 precursor miRNAs (pre-miRNAs) [8]. Pre-miRNAs comprise Phillip D. Zamore,1,* and Stefan L. Ameres1,* a single-stranded loop and a partially base-paired stem whose 1Howard Hughes Medical Institute and Department of termini bear the hallmarks of RNase III processing: a two-nucle- Biochemistry and Molecular Pharmacology otide 30 overhang, a 50 phosphate, and a 30 hydroxyl group. 2Program in Bioinformatics and Integrative Biology Nuclear pre-miRNAs are exported by Exportin 5 to the cyto- University of Massachusetts Medical School, plasm, where the RNase III enzyme Dicer liberates w22 nt 364 Plantation Street, Worcester, MA 01605, USA mature miRNA/miRNA* duplexes from the pre-miRNA stem [9–12]. Like all Dicer products, miRNA duplexes contain two- nucleotide 30 overhangs, 50 phosphate, and 30 hydroxyl groups. Summary In flies, Dicer-1 cleaves pre-miRNAs to miRNAs, whereas Dicer-2 converts long double-stranded RNA (dsRNA) into Background: MicroRNAs (miRNAs) are w22 nucleotide (nt) small interfering RNAs (siRNAs), which direct RNA interference small RNAs that control development, physiology, and pathol- (RNAi), a distinct small RNA silencing pathway required for ogy in animals and plants. Production of miRNAs involves the host defense against viral infection and somatic transposon sequential processing of primary hairpin-containing RNA poly- mobilization, as well as gene silencing triggered by exogenous merase II transcripts by the RNase III enzymes Drosha in the dsRNA [13, 14]. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Ribonuclease A
Chem. Rev. 1998, 98, 1045−1065 1045 Ribonuclease A Ronald T. Raines Departments of Biochemistry and Chemistry, University of WisconsinsMadison, Madison, Wisconsin 53706 Received October 10, 1997 (Revised Manuscript Received January 12, 1998) Contents I. Introduction 1045 II. Heterologous Production 1046 III. Structure 1046 IV. Folding and Stability 1047 A. Disulfide Bond Formation 1047 B. Prolyl Peptide Bond Isomerization 1048 V. RNA Binding 1048 A. Subsites 1048 B. Substrate Specificity 1049 C. One-Dimensional Diffusion 1049 D. Processive Catalysis 1050 VI. Substrates 1050 VII. Inhibitors 1051 Ronald T. Raines was born in 1958 in Montclair, NJ. He received Sc.B. VIII. Reaction Mechanism 1052 degrees in chemistry and biology from the Massachusetts Institute of A. His12 and His119 1053 Technology. At M.I.T., he worked with Christopher T. Walsh to reveal the reaction mechanisms of pyridoxal 5′-phosphate-dependent enzymes. B. Lys41 1054 Raines was a National Institutes of Health predoctoral fellow in the C. Asp121 1055 chemistry department at Harvard University. There, he worked with D. Gln11 1056 Jeremy R. Knowles to elucidate the reaction energetics of triosephosphate IX. Reaction Energetics 1056 isomerase. Raines was a Helen Hay Whitney postdoctoral fellow in the biochemistry and biophysics department at the University of California, A. Transphosphorylation versus Hydrolysis 1056 San Francisco. At U.C.S.F., he worked with William J. Rutter to clone, B. Rate Enhancement 1057 express, and mutate the cDNA that codes for ribonuclease A. Raines X. Ribonuclease S 1058 then joined the faculty of the biochemistry department at the University s A. S-Protein−S-Peptide Interaction 1058 of Wisconin Madison, where he is now associate professor of biochem- istry and chemistry. -

Protein Engineering to Exploit and Explore Bovine Secretory Ribonucleases
PROTEIN ENGINEERING TO EXPLOIT A N D EXPLORE BOVINE SECRETORY RIBONUCLEASES by JIN-SOO KIM A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biochemistry) at the UNIVERSITY OF WISCONSIN-MADISON 1994 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. ACKNOWLEDGEMENTS I would like to thank Dr. Ronald T. Raines for his advice and support. His scientific insight has been very helpful throughout this work. I would also like to thank the entire Raines group for their friendship and companionship. I am grateful to Dr. J. Soucek and Dr. J. Matousek for their collaboration with us, which has been a valuable part of the BS-RNase research. I thank Dr. M. Karpeisky for suggesting the protein fusion project, and Dr. G. D'Alessio and Dr. L. Mazzarella for providing the coordinates of BS-RNase. I have been generously supported by Steenbock predoctoral fellowship from the Department of Biochemistry. Finally, I thank my parents, who have encouraged (or at least not discouraged) me to pursue a career in science since I was a kid. Reproduced with permission of the copyright owner. Further reproduction prohibited without permission ABSTRACT PROTEIN ENGINEERING TO EXPLOIT AND EXPLORE BOVINE SECRETORY RIBONUCLEASES Jin-Soo Kim Under the supervision of Dr. Ronald T. Raines at the University of Wisconsin-Madison Ribonuclease S-peptide (residues 1-20) and S-protein (residues 21- 124) are the enzymatically inactive products of the limited digestion of bovine pancreatic ribonuclease A (RNase A) by subtilisin. S-Peptide binds S-protein with high affinity to form RNase S, which has full enzymatic activity. -

BRCA1 Binds TERRA RNA and Suppresses R-Loop-Based Telomeric DNA Damage ✉ Jekaterina Vohhodina 1,2 , Liana J
ARTICLE https://doi.org/10.1038/s41467-021-23716-6 OPEN BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage ✉ Jekaterina Vohhodina 1,2 , Liana J. Goehring1, Ben Liu1,2, Qing Kong1,2, Vladimir V. Botchkarev Jr.1,2, Mai Huynh1, Zhiqi Liu1, Fieda O. Abderazzaq1,2, Allison P. Clark1,2, Scott B. Ficarro1,3,4,5, Jarrod A. Marto 1,3,4,5, ✉ Elodie Hatchi 1,2 & David M. Livingston 1,2 R-loop structures act as modulators of physiological processes such as transcription termi- 1234567890():,; nation, gene regulation, and DNA repair. However, they can cause transcription-replication conflicts and give rise to genomic instability, particularly at telomeres, which are prone to forming DNA secondary structures. Here, we demonstrate that BRCA1 binds TERRA RNA, directly and physically via its N-terminal nuclear localization sequence, as well as telomere- specific shelterin proteins in an R-loop-, and a cell cycle-dependent manner. R-loop-driven BRCA1 binding to CpG-rich TERRA promoters represses TERRA transcription, prevents TERRA R-loop-associated damage, and promotes its repair, likely in association with SETX and XRN2. BRCA1 depletion upregulates TERRA expression, leading to overly abundant TERRA R-loops, telomeric replication stress, and signs of telomeric aberrancy. Moreover, BRCA1 mutations within the TERRA-binding region lead to an excess of TERRA-associated R- loops and telomeric abnormalities. Thus, normal BRCA1/TERRA binding suppresses telomere-centered genome instability. 1 Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA. 2 Department of Genetics, Harvard Medical School, Boston, MA, USA. 3 Blais Proteomics Center, Dana-Farber Cancer Institute, Boston, MA, USA. -

Role for Cytoplasmic Nucleotide Hydrolysis in Hepatic Function and Protein Synthesis
Role for cytoplasmic nucleotide hydrolysis in hepatic function and protein synthesis Benjamin H. Hudson1, Joshua P. Frederick1, Li Yin Drake, Louis C. Megosh, Ryan P. Irving, and John D. York2,3 Department of Pharmacology and Cancer Biology, Howard Hughes Medical Institute, Duke University Medical Center, Durham, NC 27710 Edited by David W. Russell, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 11, 2013 (received for review March 26, 2012) Nucleotide hydrolysis is essential for many aspects of cellular (5–7), which is degraded to 5′-AMP by a family of enzymes known function. In the case of 3′,5′-bisphosphorylated nucleotides, mam- as 3′-nucleotidases (8). mals possess two related 3′-nucleotidases, Golgi-resident 3′-phos- Mammalian genomes encode two 3′-nucleotidases, the recently phoadenosine 5′-phosphate (PAP) phosphatase (gPAPP) and characterized Golgi-resident PAP phosphatase (gPAPP) and Bisphosphate 3′-nucleotidase 1 (Bpnt1). gPAPP and Bpnt1 localize Bisphosphate 3′-nucleotidase 1 (Bpnt1), which localize to the to distinct subcellular compartments and are members of a con- Golgi lumen and cytoplasm, respectively (Fig. 1B)(8–11). gPAPP served family of metal-dependent lithium-sensitive enzymes. Al- and Bpnt1 are members of a family of small-molecule phospha- though recent studies have demonstrated the importance of tases whose activities are both dependent on divalent cations and gPAPP for proper skeletal development in mice and humans, the inhibited by lithium (8). The family comprises seven mammalian role of Bpnt1 in mammals remains largely unknown. Here we re- gene products: fructose bisphosphatase 1 and 2, inositol monophos- port that mice deficient for Bpnt1 do not exhibit skeletal defects phatase 1 and 2, inositol polyphosphate 1-phosphatase, gPAPP, but instead develop severe liver pathologies, including hypopro- and Bpnt1 (Fig. -

Insights Into the Structure and Architecture of the CCR4–NOT Complex
REVIEW ARTICLE published: 16 May 2014 doi: 10.3389/fgene.2014.00137 Insights into the structure and architecture of the CCR4–NOT complex Kun Xu 1,2 ,Yuwei Bai 1, Aili Zhang 1,2, Qionglin Zhang 2 and Mark G. Bartlam1,2* 1 State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China 2 College of Life Sciences, Nankai University, Tianjin, China Edited by: The CCR4–NOT complex is a highly conserved, multifunctional machinery with a general Martine Anne Collart, University of role in controlling mRNA metabolism. It has been implicated in a number of different Geneva, Switzerland aspects of mRNA and protein expression, including mRNA degradation, transcription Reviewed by: initiation and elongation, ubiquitination, and protein modification. The core CCR4–NOT Walter Lukiw, Louisiana State University, USA complex is evolutionarily conserved and consists of at least three NOT proteins and two Sebastiaan Winkler, University of catalytic subunits.The L-shapedcomplex is characterized by two functional modules bound Nottingham, UK to the CNOT1/Not1 scaffold protein: the deadenylase or nuclease module containing *Correspondence: two enzymes required for deadenylation, and the NOT module. In this review, we will Mark G. Bartlam, College of Life summarize the currently available information regarding the three-dimensional structure Sciences, Nankai University, 94 Weijin Road, Tianjin 300071, China and assembly of the CCR4–NOT complex, in order to provide insight into its roles in mRNA e-mail: [email protected] degradation and other biological processes. Keywords: CCR4–NOT complex, mRNA decay, poly(A), deadenylation, NOT module, protein structure INTRODUCTION 0.9–1.2 MDa in size (Liu et al., 1998). -

Signature Redacted Certified By: Ronald T
Endogenous and Chemical Modifications of Model Proteins by Valerie T. Ressler B.A. Chemistry Macalester College, 2011 M.S. Chemistry College of William & Mary, 2013 Submitted to the Department of Chemistry in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY IN CHEMISTRY at the Massachusetts Institute of Technology February 2019 @ 2019 Massachusetts Institute of Technology. All rights reserved. Signature Signature of Author: redacted____ Department of Chemistry January 14, 2019 -Signature redacted Certified by: Ronald T. Raines Firmeni Pro essor of Chemistry hesis Supervisor Signature redacted Accepted by: Robert W. Field OFTECHNOWGO Haslam and Dewey Professor of Chemistry Chairman, Departmental Committee on Graduate Students MAR 21 2019 LIBRARIES ARCHIVES 1 This doctoral thesis has been examined by a committee of professors from the Department of Chemistry as follows: Signature redacted Matthew D. Shoulders Whitehead CD Associate Professor Thesis Committee Chair Signature redacted Ronald T. Raines Firmenich Professor of Chemistry I A Thesis Supervisor Signature redacted Laura L. Kiessling Ne~ovartis Professor of Chemistry Thesis Committee Member 2 Endogenous and Chemical Modifications of Model Proteins by Valerie T. Ressler Submitted to the Department of Chemistry on January 15, 2019 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Chemistry Abstract Protein modifications are ubiquitous in nature, introducing biological complexity and functional diversity. Of the known post-translational modifications, glycosylation is one of the most common and most complex, yet some of the biological implications of this modification remain poorly understood. The development of chemical tools to mimic these modifications is helping to elucidate their biological roles and improve the range of biopharmaceuticals. -

Biochemical Characterization of Exoribonuclease Encoded by SARS Coronavirus
Journal of Biochemistry and Molecular Biology, Vol. 40, No. 5, September 2007, pp. 649-655 Biochemical Characterization of Exoribonuclease Encoded by SARS Coronavirus Ping Chen1, Miao Jiang1, Tao Hu1, Qingzhen Liu1, Xiaojiang S. Chen2 and Deyin Guo1,* 1State Key Laboratory of Virology and The Modern Virology Research Centre, College of Life Sciences, Wuhan University, Wuhan 430072, P.R. China 2Department of Molecular and Computational Biology, University of Southern California, Los Angeles, CA 90089, USA Received 2 February 2007, Accepted 6 April 2007 The nsp14 protein is an exoribonuclease that is encoded by Many published sequences of SARS-CoV have provided severe acute respiratory syndrome coronavirus (SARS- important information on the 29.7 kb positive-strand RNA CoV). We have cloned and expressed the nsp14 protein in genome of SARS-CoV (Marra et al., 2003; Rota et al., 2003; Escherichia coli, and characterized the nature and the Ruan et al., 2003). Like other coronavirus, SARS-CoV gene role(s) of the metal ions in the reaction chemistry. The expression and genome replication require polyprotein synthesis purified recombinant nsp14 protein digested a 5'-labeled and subgenomic production (Miller and Koev, 2000; Ruan et RNA molecule, but failed to digest the RNA substrate that al., 2003; Snijder et al., 2003; Thiel et al., 2003; Hussain et is modified with fluorescein group at the 3'-hydroxyl al., 2005). group, suggesting a 3'-to-5' exoribonuclease activity. The Coronavirus replication in cells is initiated by translation of exoribonuclease activity requires Mg2+ as a cofactor. the two overlapping polyproteins that are proteolytically Isothermal titration calorimetry (ITC) analysis indicated a processed to yield replication-associated proteins, including two-metal binding mode for divalent cations by nsp14. -
Generate Metabolic Map Poster
Authors: Zheng Zhao, Delft University of Technology Marcel A. van den Broek, Delft University of Technology S. Aljoscha Wahl, Delft University of Technology Wilbert H. Heijne, DSM Biotechnology Center Roel A. Bovenberg, DSM Biotechnology Center Joseph J. Heijnen, Delft University of Technology An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Marco A. van den Berg, DSM Biotechnology Center Peter J.T. Verheijen, Delft University of Technology Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. PchrCyc: Penicillium rubens Wisconsin 54-1255 Cellular Overview Connections between pathways are omitted for legibility. Liang Wu, DSM Biotechnology Center Walter M. van Gulik, Delft University of Technology L-quinate phosphate a sugar a sugar a sugar a sugar multidrug multidrug a dicarboxylate phosphate a proteinogenic 2+ 2+ + met met nicotinate Mg Mg a cation a cation K + L-fucose L-fucose L-quinate L-quinate L-quinate ammonium UDP ammonium ammonium H O pro met amino acid a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar K oxaloacetate L-carnitine L-carnitine L-carnitine 2 phosphate quinic acid brain-specific hypothetical hypothetical hypothetical hypothetical