The Rnase III Family: a Conserved Structure and Expanding Functions in Eukaryotic Dsrna Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
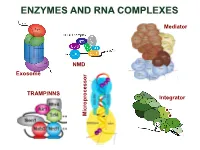
Enzymes and Rna Complexes
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Ribonuclease A
Chem. Rev. 1998, 98, 1045−1065 1045 Ribonuclease A Ronald T. Raines Departments of Biochemistry and Chemistry, University of WisconsinsMadison, Madison, Wisconsin 53706 Received October 10, 1997 (Revised Manuscript Received January 12, 1998) Contents I. Introduction 1045 II. Heterologous Production 1046 III. Structure 1046 IV. Folding and Stability 1047 A. Disulfide Bond Formation 1047 B. Prolyl Peptide Bond Isomerization 1048 V. RNA Binding 1048 A. Subsites 1048 B. Substrate Specificity 1049 C. One-Dimensional Diffusion 1049 D. Processive Catalysis 1050 VI. Substrates 1050 VII. Inhibitors 1051 Ronald T. Raines was born in 1958 in Montclair, NJ. He received Sc.B. VIII. Reaction Mechanism 1052 degrees in chemistry and biology from the Massachusetts Institute of A. His12 and His119 1053 Technology. At M.I.T., he worked with Christopher T. Walsh to reveal the reaction mechanisms of pyridoxal 5′-phosphate-dependent enzymes. B. Lys41 1054 Raines was a National Institutes of Health predoctoral fellow in the C. Asp121 1055 chemistry department at Harvard University. There, he worked with D. Gln11 1056 Jeremy R. Knowles to elucidate the reaction energetics of triosephosphate IX. Reaction Energetics 1056 isomerase. Raines was a Helen Hay Whitney postdoctoral fellow in the biochemistry and biophysics department at the University of California, A. Transphosphorylation versus Hydrolysis 1056 San Francisco. At U.C.S.F., he worked with William J. Rutter to clone, B. Rate Enhancement 1057 express, and mutate the cDNA that codes for ribonuclease A. Raines X. Ribonuclease S 1058 then joined the faculty of the biochemistry department at the University s A. S-Protein−S-Peptide Interaction 1058 of Wisconin Madison, where he is now associate professor of biochem- istry and chemistry. -

Nucleotide Sequence Surrounding a Ribonuclease III Processing Site In
Proc. Nati. Acad. Sci. USA Vol. 74, No. 3, pp. 984-988, March 1977 Biochemistry Nucleotide sequence surrounding a ribonuclease III processing site in bacteriophage T7 RNA (intercistronic region/polycistronic mRNA precursor/hairpin structure/endoribonuclease III) MARTIN ROSENBERG* AND RICHARD A. KRAMERt f * Laboratory of Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20014; and t Department of Biochemistry, Stanford University School of Medicine, Stanford, California 94305 Communicated by Charles Yanofsky, January 3, 1977 ABSTRACT We have determined a nucleotide sequence of the 5' end of the gene 0.7 mRNAs, (ii) some fragments that 87 residues surrounding a ribonuclease III (endoribonuclease contain the RNase III cleavage site are not recognized and III; EC 3.1.4.24) processing site in the bacteriophage 17 inter- cistronic region between early genes 0.3 and 0.7. The structural cleaved by the enzyme; and (iii) the 3'-terminal oligoadenylate requirements necessary for proper recognition and cleavage by sequences found on the ends of the in vdvo T7 early mRNAs (11) RNase III are discussed. In addition, other structural features are not encoded by the genome, but presumably represent a characteristic of this intercistronic boundary are described. nontemplate-dependent post-transcriptional modification. Here, we report the complete nucleotide sequence of the gene When bacteriophage T7 infects Escherichia coli, the host RNA 0.3-0.7 intercistronic region of T7 and propose a specific role polymerase (RNA nucleotidyltransferase, EC 2.7.7.6) tran- for RNA secondary structure in substrate recognition and action scribes only the early region of the phage genome (i.e., leftmost of RNase III. -

Characterization of the Mammalian RNA Exonuclease 5/NEF-Sp As a Testis-Specific Nuclear 3′′′′′ → 5′′′′′ Exoribonuclease
Downloaded from rnajournal.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Characterization of the mammalian RNA exonuclease 5/NEF-sp as a testis-specific nuclear 3′′′′′ → 5′′′′′ exoribonuclease SARA SILVA,1,2 DAVID HOMOLKA,1 and RAMESH S. PILLAI1 1Department of Molecular Biology, University of Geneva, CH-1211 Geneva 4, Switzerland 2European Molecular Biology Laboratory, Grenoble Outstation, 38042, France ABSTRACT Ribonucleases catalyze maturation of functional RNAs or mediate degradation of cellular transcripts, activities that are critical for gene expression control. Here we identify a previously uncharacterized mammalian nuclease family member NEF-sp (RNA exonuclease 5 [REXO5] or LOC81691) as a testis-specific factor. Recombinant human NEF-sp demonstrates a divalent metal ion-dependent 3′′′′′ → 5′′′′′ exoribonuclease activity. This activity is specific to single-stranded RNA substrates and is independent of their length. The presence of a 2′′′′′-O-methyl modification at the 3′′′′′ end of the RNA substrate is inhibitory. Ectopically expressed NEF-sp localizes to the nucleolar/nuclear compartment in mammalian cell cultures and this is dependent on an amino-terminal nuclear localization signal. Finally, mice lacking NEF-sp are viable and display normal fertility, likely indicating overlapping functions with other nucleases. Taken together, our study provides the first biochemical and genetic exploration of the role of the NEF-sp exoribonuclease in the mammalian genome. Keywords: NEF-sp; LOC81691; Q96IC2; REXON; RNA exonuclease 5; REXO5; 2610020H08Rik INTRODUCTION clease-mediated processing to create their final 3′ ends: poly(A) tails of most mRNAs or the hairpin structure of Spermatogenesis is the process by which sperm cells are replication-dependent histone mRNAs (Colgan and Manley produced in the male germline. -

Protein Engineering to Exploit and Explore Bovine Secretory Ribonucleases
PROTEIN ENGINEERING TO EXPLOIT A N D EXPLORE BOVINE SECRETORY RIBONUCLEASES by JIN-SOO KIM A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biochemistry) at the UNIVERSITY OF WISCONSIN-MADISON 1994 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. ACKNOWLEDGEMENTS I would like to thank Dr. Ronald T. Raines for his advice and support. His scientific insight has been very helpful throughout this work. I would also like to thank the entire Raines group for their friendship and companionship. I am grateful to Dr. J. Soucek and Dr. J. Matousek for their collaboration with us, which has been a valuable part of the BS-RNase research. I thank Dr. M. Karpeisky for suggesting the protein fusion project, and Dr. G. D'Alessio and Dr. L. Mazzarella for providing the coordinates of BS-RNase. I have been generously supported by Steenbock predoctoral fellowship from the Department of Biochemistry. Finally, I thank my parents, who have encouraged (or at least not discouraged) me to pursue a career in science since I was a kid. Reproduced with permission of the copyright owner. Further reproduction prohibited without permission ABSTRACT PROTEIN ENGINEERING TO EXPLOIT AND EXPLORE BOVINE SECRETORY RIBONUCLEASES Jin-Soo Kim Under the supervision of Dr. Ronald T. Raines at the University of Wisconsin-Madison Ribonuclease S-peptide (residues 1-20) and S-protein (residues 21- 124) are the enzymatically inactive products of the limited digestion of bovine pancreatic ribonuclease A (RNase A) by subtilisin. S-Peptide binds S-protein with high affinity to form RNase S, which has full enzymatic activity. -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900114035Cyc: Amycolatopsis sacchari DSM 44468 Cellular Overview Connections between pathways are omitted for legibility. -
Figure S1. Reverse Transcription‑Quantitative PCR Analysis of ETV5 Mrna Expression Levels in Parental and ETV5 Stable Transfectants
Figure S1. Reverse transcription‑quantitative PCR analysis of ETV5 mRNA expression levels in parental and ETV5 stable transfectants. (A) Hec1a and Hec1a‑ETV5 EC cell lines; (B) Ishikawa and Ishikawa‑ETV5 EC cell lines. **P<0.005, unpaired Student's t‑test. EC, endometrial cancer; ETV5, ETS variant transcription factor 5. Figure S2. Survival analysis of sample clusters 1‑4. Kaplan Meier graphs for (A) recurrence‑free and (B) overall survival. Survival curves were constructed using the Kaplan‑Meier method, and differences between sample cluster curves were analyzed by log‑rank test. Figure S3. ROC analysis of hub genes. For each gene, ROC curve (left) and mRNA expression levels (right) in control (n=35) and tumor (n=545) samples from The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer cohort are shown. mRNA levels are expressed as Log2(x+1), where ‘x’ is the RSEM normalized expression value. ROC, receiver operating characteristic. Table SI. Clinicopathological characteristics of the GSE17025 dataset. Characteristic n % Atrophic endometrium 12 (postmenopausal) (Control group) Tumor stage I 91 100 Histology Endometrioid adenocarcinoma 79 86.81 Papillary serous 12 13.19 Histological grade Grade 1 30 32.97 Grade 2 36 39.56 Grade 3 25 27.47 Myometrial invasiona Superficial (<50%) 67 74.44 Deep (>50%) 23 25.56 aMyometrial invasion information was available for 90 of 91 tumor samples. Table SII. Clinicopathological characteristics of The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer dataset. Characteristic n % Solid tissue normal 16 Tumor samples Stagea I 226 68.278 II 19 5.740 III 70 21.148 IV 16 4.834 Histology Endometrioid 271 81.381 Mixed 10 3.003 Serous 52 15.616 Histological grade Grade 1 78 23.423 Grade 2 91 27.327 Grade 3 164 49.249 Molecular subtypeb POLE 17 7.328 MSI 65 28.017 CN Low 90 38.793 CN High 60 25.862 CN, copy number; MSI, microsatellite instability; POLE, DNA polymerase ε. -

Monilinia Fructicola, Monilinia Laxa and Monilinia Fructigena, the Causal Agents of Brown Rot on Stone Fruits Rita M
De Miccolis Angelini et al. BMC Genomics (2018) 19:436 https://doi.org/10.1186/s12864-018-4817-4 RESEARCH ARTICLE Open Access De novo assembly and comparative transcriptome analysis of Monilinia fructicola, Monilinia laxa and Monilinia fructigena, the causal agents of brown rot on stone fruits Rita M. De Miccolis Angelini* , Domenico Abate, Caterina Rotolo, Donato Gerin, Stefania Pollastro and Francesco Faretra Abstract Background: Brown rots are important fungal diseases of stone and pome fruits. They are caused by several Monilinia species but M. fructicola, M. laxa and M. fructigena are the most common all over the world. Although they have been intensively studied, the availability of genomic and transcriptomic data in public databases is still scant. We sequenced, assembled and annotated the transcriptomes of the three pathogens using mRNA from germinating conidia and actively growing mycelia of two isolates of opposite mating types per each species for comparative transcriptome analyses. Results: Illumina sequencing was used to generate about 70 million of paired-end reads per species, that were de novo assembled in 33,861 contigs for M. fructicola, 31,103 for M. laxa and 28,890 for M. fructigena. Approximately, 50% of the assembled contigs had significant hits when blasted against the NCBI non-redundant protein database and top-hits results were represented by Botrytis cinerea, Sclerotinia sclerotiorum and Sclerotinia borealis proteins. More than 90% of the obtained sequences were complete, the percentage of duplications was always less than 14% and fragmented and missing transcripts less than 5%. Orthologous transcripts were identified by tBLASTn analysis using the B. -

Unknown Areas of Activity of Human Ribonuclease Dicer: a Putative Deoxyribonuclease Activity
molecules Article Unknown Areas of Activity of Human Ribonuclease Dicer: A Putative Deoxyribonuclease Activity Marta Wojnicka , Agnieszka Szczepanska and Anna Kurzynska-Kokorniak * Department of Ribonucleoprotein Biochemistry, Institute of Bioorganic Chemistry Polish Academy of Sciences, 61-704 Poznan, Poland; [email protected] (M.W.); [email protected] (A.S.) * Correspondence: [email protected] Received: 31 January 2020; Accepted: 17 March 2020; Published: 20 March 2020 Abstract: The Dicer ribonuclease plays a crucial role in the biogenesis of small regulatory RNAs (srRNAs) by processing long double-stranded RNAs and single-stranded hairpin RNA precursors into small interfering RNAs (siRNAs) and microRNAs (miRNAs), respectively. Dicer-generated srRNAs can control gene expression by targeting complementary transcripts and repressing their translation or inducing their cleavage. Human Dicer (hDicer) is a multidomain enzyme comprising a putative helicase domain, a DUF283 domain, platform, a PAZ domain, a connector helix, two RNase III domains (RNase IIIa and RNase IIIb) and a dsRNA-binding domain. Specific, ~20-base pair siRNA or miRNA duplexes with 2 nucleotide (nt) 3’-overhangs are generated by Dicer when an RNA substrate is anchored within the platform-PAZ-connector helix (PPC) region. However, increasing number of reports indicate that in the absence of the PAZ domain, binding of RNA substrates can occur by other Dicer domains. Interestingly, truncated variants of Dicer, lacking the PPC region, have been found to display a DNase activity. Inspired by these findings, we investigated how the lack of the PAZ domain, or the entire PPC region, would influence the cleavage activity of hDicer. Using immunopurified 3xFlag-hDicer produced in human cells and its two variants: one lacking the PAZ domain, and the other lacking the entire PPC region, we show that the PAZ domain deletion variants of hDicer are not able to process a pre-miRNA substrate, a dsRNA with 2-nt 30-overhangs, and a blunt-ended dsRNA. -

Insights Into the Structure and Architecture of the CCR4–NOT Complex
REVIEW ARTICLE published: 16 May 2014 doi: 10.3389/fgene.2014.00137 Insights into the structure and architecture of the CCR4–NOT complex Kun Xu 1,2 ,Yuwei Bai 1, Aili Zhang 1,2, Qionglin Zhang 2 and Mark G. Bartlam1,2* 1 State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China 2 College of Life Sciences, Nankai University, Tianjin, China Edited by: The CCR4–NOT complex is a highly conserved, multifunctional machinery with a general Martine Anne Collart, University of role in controlling mRNA metabolism. It has been implicated in a number of different Geneva, Switzerland aspects of mRNA and protein expression, including mRNA degradation, transcription Reviewed by: initiation and elongation, ubiquitination, and protein modification. The core CCR4–NOT Walter Lukiw, Louisiana State University, USA complex is evolutionarily conserved and consists of at least three NOT proteins and two Sebastiaan Winkler, University of catalytic subunits.The L-shapedcomplex is characterized by two functional modules bound Nottingham, UK to the CNOT1/Not1 scaffold protein: the deadenylase or nuclease module containing *Correspondence: two enzymes required for deadenylation, and the NOT module. In this review, we will Mark G. Bartlam, College of Life summarize the currently available information regarding the three-dimensional structure Sciences, Nankai University, 94 Weijin Road, Tianjin 300071, China and assembly of the CCR4–NOT complex, in order to provide insight into its roles in mRNA e-mail: [email protected] degradation and other biological processes. Keywords: CCR4–NOT complex, mRNA decay, poly(A), deadenylation, NOT module, protein structure INTRODUCTION 0.9–1.2 MDa in size (Liu et al., 1998). -

Signature Redacted Certified By: Ronald T
Endogenous and Chemical Modifications of Model Proteins by Valerie T. Ressler B.A. Chemistry Macalester College, 2011 M.S. Chemistry College of William & Mary, 2013 Submitted to the Department of Chemistry in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY IN CHEMISTRY at the Massachusetts Institute of Technology February 2019 @ 2019 Massachusetts Institute of Technology. All rights reserved. Signature Signature of Author: redacted____ Department of Chemistry January 14, 2019 -Signature redacted Certified by: Ronald T. Raines Firmeni Pro essor of Chemistry hesis Supervisor Signature redacted Accepted by: Robert W. Field OFTECHNOWGO Haslam and Dewey Professor of Chemistry Chairman, Departmental Committee on Graduate Students MAR 21 2019 LIBRARIES ARCHIVES 1 This doctoral thesis has been examined by a committee of professors from the Department of Chemistry as follows: Signature redacted Matthew D. Shoulders Whitehead CD Associate Professor Thesis Committee Chair Signature redacted Ronald T. Raines Firmenich Professor of Chemistry I A Thesis Supervisor Signature redacted Laura L. Kiessling Ne~ovartis Professor of Chemistry Thesis Committee Member 2 Endogenous and Chemical Modifications of Model Proteins by Valerie T. Ressler Submitted to the Department of Chemistry on January 15, 2019 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Chemistry Abstract Protein modifications are ubiquitous in nature, introducing biological complexity and functional diversity. Of the known post-translational modifications, glycosylation is one of the most common and most complex, yet some of the biological implications of this modification remain poorly understood. The development of chemical tools to mimic these modifications is helping to elucidate their biological roles and improve the range of biopharmaceuticals.