Babela Massiliensis, a Representative of a Widespread Bacterial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
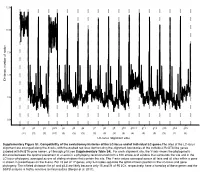
LC-Locus Alignment Sites Distance, Number of Nodes Supplementary
12.5 10.0 7.5 5.0 Distance, number of nodes 2.5 0.0 g1 g2 g3 g3.5 g4 g5 g6 g7 g8 g9 g10 g10.1 g11 g12 g13 g14 g15 (11) (3) (3) (10) (3) (3) (3) (3) (3) (3) (3) (4) (3) (3) (3) (2) (3) LC-locus alignment sites Supplementary Figure S1. Compatibility of the evolutionary histories of the LC-locus and of individual LC genes.The sites of the LC-locus alignment are arranged along the X-axis, with the dashed red lines demarcating the alignment boundaries of the individual RcGTA-like genes (labeled with RcGTA gene names, g1 through g15; see Supplementary Table S4). For each alignment site, the Y-axis shows the phylogenetic distance between the optimal placement of a taxon in a phylogeny reconstructed from a 100 amino-acid window that surrounds the site and in the LC-locus phylogeny, averaged across all sliding windows that contain the site. The Y-axis values averaged across all taxa and all sites within a gene is shown in parentheses on the X-axis. For 15 out of 17 genes, only 2-4 nodes separate the optimal taxon position in the LC-locus and gene phylogeny. The inflated distances for g1 and g3.5 are likely because only 15 and 21 of 95 LCs, respectively, have a homolog of these genes and the SSPB analysis is highly sensitive to missing data (Berger et al. 2011). a. Bacteria Unassigned Thermotogae Tenericutes Synergistetes Spirochaetes Proteobacteria ylum Planctomycetes h p Firmicutes Deferribacteres Cyanobacteria Chloroflexi Bacteroidetes Actinobacteria Acidobacteria 1(11,750) 2(1,750) 3(2,538) 4(168) 5(51) 6(54) 7(43) 8(32) 9(26) 10(40) 11(33) 12(198) 13(173) 14(101) 15(98) 16(43) 17(114) Number of rcc01682−rcc01698 homologs in a cluster b. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup -

Gene Gain and Loss Events in Rickettsia and Orientia Species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2*
Georgiades et al. Biology Direct 2011, 6:6 http://www.biology-direct.com/content/6/1/6 RESEARCH Open Access Gene gain and loss events in Rickettsia and Orientia species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2* Abstract Background: Genome degradation is an ongoing process in all members of the Rickettsiales order, which makes these bacterial species an excellent model for studying reductive evolution through interspecies variation in genome size and gene content. In this study, we evaluated the degree to which gene loss shaped the content of some Rickettsiales genomes. We shed light on the role played by horizontal gene transfers in the genome evolution of Rickettsiales. Results: Our phylogenomic tree, based on whole-genome content, presented a topology distinct from that of the whole core gene concatenated phylogenetic tree, suggesting that the gene repertoires involved have different evolutionary histories. Indeed, we present evidence for 3 possible horizontal gene transfer events from various organisms to Orientia and 6 to Rickettsia spp., while we also identified 3 possible horizontal gene transfer events from Rickettsia and Orientia to other bacteria. We found 17 putative genes in Rickettsia spp. that are probably the result of de novo gene creation; 2 of these genes appear to be functional. On the basis of these results, we were able to reconstruct the gene repertoires of “proto-Rickettsiales” and “proto-Rickettsiaceae”, which correspond to the ancestors of Rickettsiales and Rickettsiaceae, respectively. Finally, we found that 2,135 genes were lost during the evolution of the Rickettsiaceae to an intracellular lifestyle. Conclusions: Our phylogenetic analysis allowed us to track the gene gain and loss events occurring in bacterial genomes during their evolution from a free-living to an intracellular lifestyle. -

High Prevalence of Rickettsia Africae Variants in Amblyomma Variegatum Ticks from Domestic Mammals in Rural Western Kenya: Implications for Human Health
n Maina, A. N. et al. (2014) High prevalence of Rickettsia africae variants in Amblyomma variegatum ticks from domestic mammals in rural western Kenya: implications for human health. Vector-Borne and Zoonotic Diseases, 14 (10). pp. 693-702. ISSN 1530-3667 Copyright © 2014 The Authors http://eprints.gla.ac.uk/99480/ Deposited on: 17 November 2014 Enlighten – Research publications by members of the University of Glasgow http://eprints.gla.ac.uk VECTOR-BORNE AND ZOONOTIC DISEASES Volume 14, Number 10, 2014 ORIGINAL ARTICLES ª Mary Ann Liebert, Inc. DOI: 10.1089/vbz.2014.1578 High Prevalence of Rickettsia africae Variants in Amblyomma variegatum Ticks from Domestic Mammals in Rural Western Kenya: Implications for Human Health Alice N. Maina,1–3 Ju Jiang,3 Sylvia A. Omulo,2 Sally J. Cutler,4 Fredrick Ade,2 Eric Ogola,2 Daniel R. Feikin,5 M. Kariuki Njenga,5 Sarah Cleaveland,6 Solomon Mpoke,1,2 Zipporah Ng’ang’a,1 Robert F. Breiman,5 Darryn L. Knobel,2,7 and Allen L. Richards3 Abstract Tick-borne spotted fever group (SFG) rickettsioses are emerging human diseases caused by obligate intracel- lular Gram-negative bacteria of the genus Rickettsia. Despite being important causes of systemic febrile illnesses in travelers returning from sub-Saharan Africa, little is known about the reservoir hosts of these pathogens. We conducted surveys for rickettsiae in domestic animals and ticks in a rural setting in western Kenya. Of the 100 serum specimens tested from each species of domestic ruminant 43% of goats, 23% of sheep, and 1% of cattle had immunoglobulin G (IgG) antibodies to the SFG rickettsiae. -

Tick-Borne Disease Working Group 2020 Report to Congress
2nd Report Supported by the U.S. Department of Health and Human Services • Office of the Assistant Secretary for Health Tick-Borne Disease Working Group 2020 Report to Congress Information and opinions in this report do not necessarily reflect the opinions of each member of the Working Group, the U.S. Department of Health and Human Services, or any other component of the Federal government. Table of Contents Executive Summary . .1 Chapter 4: Clinical Manifestations, Appendices . 114 Diagnosis, and Diagnostics . 28 Chapter 1: Background . 4 Appendix A. Tick-Borne Disease Congressional Action ................. 8 Chapter 5: Causes, Pathogenesis, Working Group .....................114 and Pathophysiology . 44 The Tick-Borne Disease Working Group . 8 Appendix B. Tick-Borne Disease Working Chapter 6: Treatment . 51 Group Subcommittees ...............117 Second Report: Focus and Structure . 8 Chapter 7: Clinician and Public Appendix C. Acronyms and Abbreviations 126 Chapter 2: Methods of the Education, Patient Access Working Group . .10 to Care . 59 Appendix D. 21st Century Cures Act ...128 Topic Development Briefs ............ 10 Chapter 8: Epidemiology and Appendix E. Working Group Charter. .131 Surveillance . 84 Subcommittees ..................... 10 Chapter 9: Federal Inventory . 93 Appendix F. Federal Inventory Survey . 136 Federal Inventory ....................11 Chapter 10: Public Input . 98 Appendix G. References .............149 Minority Responses ................. 13 Chapter 11: Looking Forward . .103 Chapter 3: Tick Biology, Conclusion . 112 Ecology, and Control . .14 Contributions U.S. Department of Health and Human Services James J. Berger, MS, MT(ASCP), SBB B. Kaye Hayes, MPA Working Group Members David Hughes Walker, MD (Co-Chair) Adalbeto Pérez de León, DVM, MS, PhD Leigh Ann Soltysiak, MS (Co-Chair) Kevin R. -

Wholly Rickettsia! Reconstructed Metabolic Profile of the Quintessential Bacterial Parasite of Eukaryotic Cells
Faculty Scholarship 2017 Wholly Rickettsia! Reconstructed Metabolic Profile of the Quintessential Bacterial Parasite of Eukaryotic Cells Timothy P. Driscoll Victoria I. Verhoeve Mark L. Guillotte Stephanie S. Lehman Sherri A. Rennoll See next page for additional authors Follow this and additional works at: https://researchrepository.wvu.edu/faculty_publications Authors Timothy P. Driscoll, Victoria I. Verhoeve, Mark L. Guillotte, Stephanie S. Lehman, Sherri A. Rennoll, Magda Beier-Sexton, M Sayeedur Rahman, Abdu F. Azad, and Joseph J. Gillespie RESEARCH ARTICLE crossm Wholly Rickettsia! Reconstructed Metabolic Profile of the Quintessential Bacterial Parasite of Eukaryotic Cells Timothy P. Driscoll,a Victoria I. Verhoeve,a Mark L. Guillotte,b Stephanie S. Lehman,b Sherri A. Rennoll,b Magda Beier-Sexton,b Downloaded from M. Sayeedur Rahman,b Abdu F. Azad,b Joseph J. Gillespieb Department of Biology, West Virginia University, Morgantown, West Virginia, USAa; Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, Maryland, USAb ABSTRACT Reductive genome evolution has purged many metabolic pathways Received 21 May 2017 Accepted 15 August from obligate intracellular Rickettsia (Alphaproteobacteria; Rickettsiaceae). While some 2017 Published 26 September 2017 aspects of host-dependent rickettsial metabolism have been characterized, the array Citation Driscoll TP, Verhoeve VI, Guillotte ML, of host-acquired metabolites and their cognate transporters remains unknown. This Lehman SS, Rennoll SA, Beier-Sexton M, http://mbio.asm.org/ Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly dearth of information has thwarted efforts to obtain an axenic Rickettsia culture, Rickettsia! Reconstructed metabolic profile of a major impediment to conventional genetic approaches. Using phylogenomics the quintessential bacterial parasite of and computational pathway analysis, we reconstructed the Rickettsia metabolic eukaryotic cells. -

Laboratory Diagnostics of Rickettsia Infections in Denmark 2008–2015
biology Article Laboratory Diagnostics of Rickettsia Infections in Denmark 2008–2015 Susanne Schjørring 1,2, Martin Tugwell Jepsen 1,3, Camilla Adler Sørensen 3,4, Palle Valentiner-Branth 5, Bjørn Kantsø 4, Randi Føns Petersen 1,4 , Ole Skovgaard 6,* and Karen A. Krogfelt 1,3,4,6,* 1 Department of Bacteria, Parasites and Fungi, Statens Serum Institut (SSI), 2300 Copenhagen, Denmark; [email protected] (S.S.); [email protected] (M.T.J.); [email protected] (R.F.P.) 2 European Program for Public Health Microbiology Training (EUPHEM), European Centre for Disease Prevention and Control (ECDC), 27180 Solnar, Sweden 3 Scandtick Innovation, Project Group, InterReg, 551 11 Jönköping, Sweden; [email protected] 4 Virus and Microbiological Special Diagnostics, Statens Serum Institut (SSI), 2300 Copenhagen, Denmark; [email protected] 5 Department of Infectious Disease Epidemiology and Prevention, Statens Serum Institut (SSI), 2300 Copenhagen, Denmark; [email protected] 6 Department of Science and Environment, Roskilde University, 4000 Roskilde, Denmark * Correspondence: [email protected] (O.S.); [email protected] (K.A.K.) Received: 19 May 2020; Accepted: 15 June 2020; Published: 19 June 2020 Abstract: Rickettsiosis is a vector-borne disease caused by bacterial species in the genus Rickettsia. Ticks in Scandinavia are reported to be infected with Rickettsia, yet only a few Scandinavian human cases are described, and rickettsiosis is poorly understood. The aim of this study was to determine the prevalence of rickettsiosis in Denmark based on laboratory findings. We found that in the Danish individuals who tested positive for Rickettsia by serology, the majority (86%; 484/561) of the infections belonged to the spotted fever group. -

Exploration of Tick-Borne Pathogens and Microbiota of Dog Ticks Collected at Potchefstroom Animal Welfare Society
Exploration of tick-borne pathogens and microbiota of dog ticks collected at Potchefstroom Animal Welfare Society C Van Wyk orcid.org 0000-0002-5971-4396 Dissertation submitted in fulfilment of the requirements for the degree Master of Science in Environmental Sciences at the North-West University Supervisor: Prof MMO Thekisoe Co-supervisor: Ms K Mtshali Graduation May 2019 24263524 DEDICATION This thesis is dedicated to the late Nettie Coetzee. For her inspiration and lessons to overcome any obstacle that life may present. God called home another angel we all love and miss you. “We are the scientists, trying to make sense of the stars inside us.” -Christopher Poindexter i ACKNOWLEDGEMENTS My sincerest appreciation goes out to my supervisor, Prof. Oriel M.M. Thekisoe, for his support, motivation, guidance, and insightfulness during the duration of this project and been there every step of the way. I would also like to thank my co-supervisor, Ms. Khethiwe Mtshali, for her patience and insightfulness towards the corrections of this thesis. I would like to thank Dr. Stalone Terera and the staff members at PAWS for their aid towards the collection of tick specimens. For the sequencing on the Illumina MiSeq platform and metagenomic data analysis I would like to thank Dr. Moeti O. Taioe, Dr. Charlotte M.S. Mienie, Dr. Danie C. La Grange, and Dr. Marlin J. Mert. I would like to thank the National Research Foundation (NRF) for their financial support by awarding me the S&F- Innovation Masters Scholarship and the North-West University (NWU) for the use of their laboratories. -

Detecting Phylogenetic Signals from Deep Roots of the Tree of Life
UNIVERSITY OF CALIFORNIA,MERCED Detecting Phylogenetic Signals From Deep Roots of the Tree of Life A dissertation submitted in partial fulfillment of the requirements for the degree Doctor of Philosophy in Quantitative and Systems Biology by Katherine Colleen Harris Amrine Committee in charge: Professor Carolin Frank, Chair Professor David Ardell Professor Meng-Lin Tsao Professor Suzanne Sindi August 2013 Copyright Katherine C. Amrine All Rights Reserved UNIVERSITY OF CALIFORNIA,MERCED Graduate Division The Dissertation of Katherine Colleen Harris Amrine is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Faculty Advisor: David H. Ardell Committee Members: Chair: Carolin Frank Meng-Lin Tsao Suzanne Sindi Date iii Contents List of Figures ................................................................. vi List of Tables .................................................................. ix Acknowledgements ............................................................. x Vita ........................................................................... xi Abstract ...................................................................... xii 1 Shifting focus in evolutionary biology – identifying a new signal for phylogenetic tree reconstruction and taxonomic classification 1 1.1 The evolution of bacterial classification and phylogeny . .1 1.2 The historical marker – 16S . .2 1.3 Complications in bacterial classification and phylogeny . .2 1.3.1 Horizontal gene transfer . .2 1.3.2 Does a true tree exist? . .3 1.4 Methods for phylogenetic tree reconstruction . .3 1.4.1 DNA . .3 1.4.2 RNA . .4 1.4.3 Proteins . .4 1.4.4 Data compilation . .5 1.5 Bias in tree-building . .5 1.6 Biological bias in biological data . .6 1.7 The tRNA interaction network . .6 1.8 Information theory . .8 1.9 Machine Learning for bacterial classification . .9 2 tRNA signatures reveal polyphyletic origins of streamlined SAR11 genomes among the Alphaproteobacteria 12 2.1 Abstract . -

WO 2013/185113 Al 12 December 2013 (12.12.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/185113 Al 12 December 2013 (12.12.2013) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 38/02 (2006.01) A61P 7/02 (2006.01) kind of national protection available): AE, AG, AL, AM, A61K 38/36 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (21) Number: International Application DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/US20 13/044841 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, (22) International Filing Date: KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 7 June 2013 (07.06.2013) MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, (25) Filing Language: English SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (26) Publication Language: English TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/657,688 8 June 2012 (08.06.2012) kind of regional protection available): ARIPO (BW, GH, 61/800,626 15 March 2013 (15.03.2013) GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, (71) Applicant: BIOGEN IDEC MA INC. -

Next Generation Sequencing Identifies Five Major Classes of Potentially Therapeutic Enzymes Secreted by Lucilia Sericata Medical Maggots
Hindawi Publishing Corporation BioMed Research International Volume 2016, Article ID 8285428, 27 pages http://dx.doi.org/10.1155/2016/8285428 Research Article Next Generation Sequencing Identifies Five Major Classes of Potentially Therapeutic Enzymes Secreted by Lucilia sericata Medical Maggots Zdenjk Franta,1 Heiko Vogel,2 Rüdiger Lehmann,1 Oliver Rupp,3 Alexander Goesmann,3 and Andreas Vilcinskas1,4 1 Department of Bioresources, Fraunhofer Institute for Molecular Biology and Applied Ecology, Winchesterstraße 2, 35394 Giessen, Germany 2Department of Entomology, Max Planck Institute for Chemical Ecology, Hans-Knoll-Straße¨ 8, 07745 Jena, Germany 3Justus-Liebig-University of Giessen, Bioinformatics and System Biology, Heinrich-Buff-Ring 58, 35392 Giessen, Germany 4Justus-Liebig-University of Giessen, Institute for Insect Biotechnology, Heinrich-Buff-Ring 26-32, 35392 Giessen, Germany Correspondence should be addressed to Andreas Vilcinskas; [email protected] Received 29 January 2016; Accepted 7 March 2016 Academic Editor: Yudong Cai Copyright © 2016 Zdenekˇ Franta et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Lucilia sericata larvae are used as an alternative treatment for recalcitrant and chronic wounds. Their excretions/secretions contain molecules that facilitate tissue debridement, disinfect, or accelerate wound healing and have therefore -

Bacteriology
SECTION 1 High Yield Microbiology 1 Bacteriology MORGAN A. PENCE Definitions Obligate/strict anaerobe: an organism that grows only in the absence of oxygen (e.g., Bacteroides fragilis). Spirochete Aerobe: an organism that lives and grows in the presence : spiral-shaped bacterium; neither gram-positive of oxygen. nor gram-negative. Aerotolerant anaerobe: an organism that shows signifi- cantly better growth in the absence of oxygen but may Gram Stain show limited growth in the presence of oxygen (e.g., • Principal stain used in bacteriology. Clostridium tertium, many Actinomyces spp.). • Distinguishes gram-positive bacteria from gram-negative Anaerobe : an organism that can live in the absence of oxy- bacteria. gen. Bacillus/bacilli: rod-shaped bacteria (e.g., gram-negative Method bacilli); not to be confused with the genus Bacillus. • A portion of a specimen or bacterial growth is applied to Coccus/cocci: spherical/round bacteria. a slide and dried. Coryneform: “club-shaped” or resembling Chinese letters; • Specimen is fixed to slide by methanol (preferred) or heat description of a Gram stain morphology consistent with (can distort morphology). Corynebacterium and related genera. • Crystal violet is added to the slide. Diphtheroid: clinical microbiology-speak for coryneform • Iodine is added and forms a complex with crystal violet gram-positive rods (Corynebacterium and related genera). that binds to the thick peptidoglycan layer of gram-posi- Gram-negative: bacteria that do not retain the purple color tive cell walls. of the crystal violet in the Gram stain due to the presence • Acetone-alcohol solution is added, which washes away of a thin peptidoglycan cell wall; gram-negative bacteria the crystal violet–iodine complexes in gram-negative appear pink due to the safranin counter stain.