CGC 15 (1992) Cucurbit Genetics Cooperative
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
THE EVOLUTION of SEED MORPHOLOGY in DOMESTICATED Chenopodium: an ARCHAEOLOGICAL CASE STUDY
]. Ethnobiol. 13(2):149-169 Winter 1993 THE EVOLUTION OF SEED MORPHOLOGY IN DOMESTICATED Chenopodium: AN ARCHAEOLOGICAL CASE STUDY KRISTEN }. GREMILLION Department of Anthropology The Ohio Stute University Columbus, OH 43210-1364 ABSTRACf.-A large body of data on several key morphological characters has been compiled through examination of collections of archaeological Chenopodium from eastern North America. Contrary to expectations based on change in certain other seed crops, the patterns of variation observed in Chenopodium do not reflect a gradual evolution of seed morphology away from the wild type. Evidence for decreasing levels of morphological variability in the evolving domesticate is like wise minimal. These findings demonstrate that the rate and character of crop evolution as revealed in the archaeological record can be expected to vary consid erably among taxa. RESUMEN.-Se ha compilado un extenso ouerpo de datos sobre varios carac teres morfol6gicos clave mediante el examen de colecciones de Chenopodium arqueol6gico del este de Norteamerica. Contrariamente a las expectativas basadas en el cambio en ciertos otms cultivos de semilla, los patrones de variaci6n obser vados en Chenopodium no reflejan una evoluci6n gradual de la morfologia de las semillas en credente distancia del tipo silvestre. La evidencia de niveles decre cientes de variabilidad morfol6gica en la especie domesticada en evoluci6n es asimismo minima. Estos resultados demuestran que puede esperarse que la 13sa y el caracter de la evoluci6n de los cultivos, tal y como se revela en el registro arqueol6gico, varien considerablemente entre taxa distintos. REsUME.-Un large ensemble de donnees concernant plusieurs characteres mor phologiques importants a ete recueilli en examinant des collections de Cheno podium de I'est de I' Amerique. -

Correlating Sensory Attributes, Textural Parameters and Volatile Organic
CORRELATING SENSORY ATTRIBUTES, TEXTURAL PARAMETERS AND VOLATILE ORGANIC COMPOUNDS FOR THE ASSESSMENT OF DISTINCTIVE QUALITY TRAITS OF MELON AND PEACH FRUIT CULTIVARS Tiago Luís Cardoso Ferreira Pinhanços de Bianchi Per citar o enllaçar aquest document: Para citar o enlazar este documento: Use this url to cite or link to this publication: http://hdl.handle.net/10803/671744 ADVERTIMENT. L'accés als continguts d'aquesta tesi doctoral i la seva utilització ha de respectar els drets de la persona autora. Pot ser utilitzada per a consulta o estudi personal, així com en activitats o materials d'investigació i docència en els termes establerts a l'art. 32 del Text Refós de la Llei de Propietat Intel·lectual (RDL 1/1996). Per altres utilitzacions es requereix l'autorització prèvia i expressa de la persona autora. En qualsevol cas, en la utilització dels seus continguts caldrà indicar de forma clara el nom i cognoms de la persona autora i el títol de la tesi doctoral. No s'autoritza la seva reproducció o altres formes d'explotació efectuades amb finalitats de lucre ni la seva comunicació pública des d'un lloc aliè al servei TDX. Tampoc s'autoritza la presentació del seu contingut en una finestra o marc aliè a TDX (framing). Aquesta reserva de drets afecta tant als continguts de la tesi com als seus resums i índexs. ADVERTENCIA. El acceso a los contenidos de esta tesis doctoral y su utilización debe respetar los derechos de la persona autora. Puede ser utilizada para consulta o estudio personal, así como en actividades o materiales de investigación y docencia en los términos establecidos en el art. -

High Tunnel Melon and Watermelon Production
High Tunnel Melon and Watermelon Production University of Missouri Extension M173 Contents Author Botany 1 Lewis W. Jett, Division of Plant Sciences, University of Missouri-Columbia Cultivar selection 3 Editorial staff Transplant production 4 MU Extension and Agricultural Information Planting in the high tunnel 5 Dale Langford, editor Dennis Murphy, illustrator Row covers 6 On the World Wide Web Soil management and fertilization 6 Find this and other MU Extension publications on the Irrigation 7 Web at http://muextension.missouri.edu Pollination 7 Photographs Pruning 8 Except where noted, photographs are by Lewis W. Jett. Trellising 8 Harvest and yield 9 Marketing 10 Pest management 10 Useful references 14 Melon and watermelon seed sources 15 Sources of high tunnels (hoophouses) 16 For further information, address questions to College of Dr. Lewis W. Jett Agriculture Extension State Vegetable Crops Specialist Food and Natural Division of Plant Sciences Resources University of Missouri Columbia, MO 65211 Copyright 2006 by the University of Missouri Board of Curators E-mail: [email protected] College of Agriculture, Food and Natural Resources High Tunnel Melon and Watermelon Production igh tunnels are low-cost, passive, melo has several botanical subgroups (Table 1). solar greenhouses that use no fossil In the United States, reticulatus and inodorus are Hfuels for heating or venting (Figure commercially grown, while the remaining groups 1). High tunnels can provide many benefits to are grown for niche or local markets. horticulture crop producers: The cantaloupe fruit that most Americans • High tunnels are used to lengthen the are familiar with is not actually a true cantaloupe. -

Climate Change and Cultural Response in the Prehistoric American Southwest
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- Published Research US Geological Survey Fall 2009 Climate Change and Cultural Response In The Prehistoric American Southwest Larry Benson U.S. Geological Survey, [email protected] Michael S. Berry Bureau of Reclamation Follow this and additional works at: https://digitalcommons.unl.edu/usgsstaffpub Benson, Larry and Berry, Michael S., "Climate Change and Cultural Response In The Prehistoric American Southwest" (2009). USGS Staff -- Published Research. 725. https://digitalcommons.unl.edu/usgsstaffpub/725 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- Published Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. CLIMATE CHANGE AND CULTURAL RESPONSE IN THE PREHISTORIC AMERICAN SOUTHWEST Larry V. Benson and Michael S. Berry ABSTRACT Comparison of regional tree-ring cutting-date distributions from the southern Col- orado Plateau and the Rio Grande region with tree-ring-based reconstructions of the Palmer Drought Severity Index (PDSI) and with the timing of archaeological stage transitions indicates that Southwestern Native American cultures were peri- odically impacted by major climatic oscillations between A.D. 860 and 1600. Site- specifi c information indicates that aggregation, abandonment, and out-migration from many archaeological regions occurred during several widespread mega- droughts, including the well-documented middle-twelfth- and late-thirteenth- century droughts. We suggest that the demographic response of southwestern Native Americans to climate variability primarily refl ects their dependence on an inordinately maize-based subsistence regimen within a region in which agricul- ture was highly sensitive to climate change. -

Sieving Methodology for Recovery of Large Cultigen Pollen
An improved methodology for the recovery of Zea mays and other large crop pollen, with implications for environmental archaeology in the Neotropics Article Accepted Version Whitney, B. S., Rushton, E. A. C., Carson, J. F., Iriarte, J. and Mayle, F. E. (2012) An improved methodology for the recovery of Zea mays and other large crop pollen, with implications for environmental archaeology in the Neotropics. The Holocene, 22 (10). pp. 1087-1096. ISSN 0959-6836 doi: https://doi.org/10.1177/0959683612441842 Available at http://centaur.reading.ac.uk/32925/ It is advisable to refer to the publisher’s version if you intend to cite from the work. See Guidance on citing . To link to this article DOI: http://dx.doi.org/10.1177/0959683612441842 Publisher: Sage Publications All outputs in CentAUR are protected by Intellectual Property Rights law, including copyright law. Copyright and IPR is retained by the creators or other copyright holders. Terms and conditions for use of this material are defined in the End User Agreement . www.reading.ac.uk/centaur CentAUR Central Archive at the University of Reading Reading’s research outputs online An improved methodology for the recovery of Zea mays and other large crop pollen, with implications for environmental archaeology in the Neotropics Bronwen S. Whitney1*, Elizabeth A. C. Rushton2, John F. Carson3,1, Jose Iriarte4, and Francis E. Mayle3 1School of Geosciences, The University of Edinburgh, Drummond St., Edinburgh EH8 9XP, UK 2School of Geography, The University of Nottingham, University Park, Nottingham NG7 2RD, UK 3School of Archaeology, Geography and Environmental Science, The University of Reading, Whiteknights, Reading RG6 6AB, UK 4Department of Archaeology, College of Humanities, University of Exeter, Exeter EX4 4QE, UK *Corresponding author, email: [email protected] tel: +44(0)131 650 9140 fax: +44(0)131 650 2524 A. -

Generate Evaluation Form
Great Lakes Fruit, Vegetable & Farm Market EXPO Michigan Greenhouse Growers EXPO December 5-7, 2017 DeVos Place Convention Center, Grand Rapids, MI Vine Crops Where: Grand Gallery (main level) Room E & F MI Recertification credits: 2 (1B, COMM CORE, PRIV CORE) OH Recertification credits: 1 (presentations as marked) CCA Credits: PM(1.0) CM(1.0) Moderator: Ben Phillips, Vegetable Extension Educator, MSU Extension, Saginaw, MI 9:00 am Integrated Management of Powdery Mildew and Gummy Stem Blight on Cucurbits: A Florida Perspective (OH: 2B, 0.5 hr) Gary Vallad, Plant Pathology Dept., Univ. of Florida 9:30 am Perspectives and Opportunities for Growing Orange Flesh and Specialty Melons Jonathan Schultheis, Dept. Horticultural Science, North Carolina State University 10:00 am Best Management Practices: Controlling Insects While Protecting Pollinators (OH: CORE, 0.5 hr) Rick Foster, Entomology Dept., Purdue Univ. 10:30 am Cover Cropping in Melons for Reduced Washing After Harvest Chris Gunter, Horticultural Science Dept., North Carolina State Univ. 11:00 am Session Ends Perspectives and Opportunities for Growing Orange Flesh and Specialty Melons Jonathan R. Schultheis North Carolina State University Department of Horticultural Science 2721 Founders Drive, 264 Kilgore Raleigh, NC 27695-7609 [email protected] Orange Flesh Melon Types: The landscape of orange flesh muskmelons (cantaloupes) has changed dramatically over the past five to ten years. Traditionally, there were two primary orange flesh melon types grown and sold; western types, which are fruits that range in size from about 3 to 5 pounds which are heavily netted and not sutured, and eastern types which have some netting, have a shorter shelf life but have a larger fruit size than a western melon, and tend to have a softer flesh with apparent more flavor than a western melon. -

Archaeological Evidence of Aboriginal Cultigen Use in Late Nineteenth and Early Twentieth Century Death Valley, California
Journal of Ethnobiology 17(2):267-282 Winter 1997 ARCHAEOLOGICAL EVIDENCE OF ABORIGINAL CULTIGEN USE IN LATE NINETEENTH AND EARLY TWENTIETH CENTURY DEATH VALLEY, CALIFORNIA ROBERT M. YOHE, II Archaeological Survey of Idaho Idaho State Historical Society Boise, Idaho 83702 ABSTRACT.-During archaeological test excavations in two rockshelters in Death Valley, California, two storage features were unearthed which were found to contain numerous perishable artifacts and foodstuffs. In addition to seed remains of indigenous species, including mesquite and pinon, several seeds of introduced cultigens were recovered from within the features, including melon, squash, and beans. The feature containing the greatest number of domesticate seeds appears to date to the late nineteenth and/or early twentieth century and represents the first reported archaeological evidence of Shoshoni horticulture in the southwestem Great Basin. RESUMEN.-Durante excavaciones arqueologicas preliminares en dos refugios de roc a en el Valle de la Muerte, en California, se descubrieron dos almacenamientos que resultaron contener numerosos artefactos y alimentos perecederos. Adernas de restos de semillas de especies nativas, incluyendo mezquite y pinon, se encontraron dentro de los vestigios varias sernillas de cultivos introducidos, incluyendo melon, calabaza y frijol. EIalmacenamiento que contenia el mayor ruimero de semillas domesticadas parece datar de finales del siglo diecinueve y/ 0 principios del siglo veinte, y representa la primera evidencia arqueologica reportada de horticultura shoshoni en el suroeste de la Gran Cuenca. RESUME.-Des reconnaissances archeologiques conduites dans deux abris rocheux de la Vallee de la Mort en Californie ont permis de mettre au jour deux structures d'entreposage contenant plusieurs objets et denrees perissables. -

Transformation of 'Galia' Melon to Improve Fruit
TRANSFORMATION OF ‘GALIA’ MELON TO IMPROVE FRUIT QUALITY By HECTOR GORDON NUÑEZ-PALENIUS A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Hector Gordon Nuñez-Palenius This document is dedicated to the seven reasons in my life, who make me wake up early all mornings, work hard in order to achieve my objectives, dream on new horizons and goals, feel the beaty of the wind, rain and sunset, but mostly because they make me believe in God: my Dads Jose Nuñez Vargas and Salvador Federico Nuñez Palenius, my Moms Janette Ann Palenius Alberi and Consuelo Nuñez Solís, my wife Nélida Contreras Sánchez, my son Hector Manuel Nuñez Contreras and my daughter Consuelo Janette Nuñez Contreras. ACKNOWLEDGMENTS This dissertation could not have been completed without the support and help of many people who are gratefully acknowledged here. My greatest debt is to Dr. Daniel James Cantliffe, who has been a dedicated advisor and mentor, but mostly an excellent friend. He provided constant and efficient guidance to my academic work and research projects. This dissertation goal would not be possible without his insightful, invariable and constructive criticism. I thank Dr. Daniel J. Cantliffe for his exceptional course on Advanced Vegetable Production Techniques (HOS-5565) and the economic support for living expenses during my graduate education in UF. I extend my appreciation to my supervisory committee, Dr. Donald J. Huber, Dr. Harry J. Klee, and Dr. Donald Hopkins, for their academic guidance. -

Dictionary of Cultivated Plants and Their Regions of Diversity Second Edition Revised Of: A.C
Dictionary of cultivated plants and their regions of diversity Second edition revised of: A.C. Zeven and P.M. Zhukovsky, 1975, Dictionary of cultivated plants and their centres of diversity 'N -'\:K 1~ Li Dictionary of cultivated plants and their regions of diversity Excluding most ornamentals, forest trees and lower plants A.C. Zeven andJ.M.J, de Wet K pudoc Centre for Agricultural Publishing and Documentation Wageningen - 1982 ~T—^/-/- /+<>?- •/ CIP-GEGEVENS Zeven, A.C. Dictionary ofcultivate d plants andthei rregion so f diversity: excluding mostornamentals ,fores t treesan d lowerplant s/ A.C .Zeve n andJ.M.J ,d eWet .- Wageninge n : Pudoc. -11 1 Herz,uitg . van:Dictionar y of cultivatedplant s andthei r centreso fdiversit y /A.C .Zeve n andP.M . Zhukovsky, 1975.- Me t index,lit .opg . ISBN 90-220-0785-5 SISO63 2UD C63 3 Trefw.:plantenteelt . ISBN 90-220-0785-5 ©Centre forAgricultura l Publishing and Documentation, Wageningen,1982 . Nopar t of thisboo k mayb e reproduced andpublishe d in any form,b y print, photoprint,microfil m or any othermean swithou t written permission from thepublisher . Contents Preface 7 History of thewor k 8 Origins of agriculture anddomesticatio n ofplant s Cradles of agriculture and regions of diversity 21 1 Chinese-Japanese Region 32 2 Indochinese-IndonesianRegio n 48 3 Australian Region 65 4 Hindustani Region 70 5 Central AsianRegio n 81 6 NearEaster n Region 87 7 Mediterranean Region 103 8 African Region 121 9 European-Siberian Region 148 10 South American Region 164 11 CentralAmerica n andMexica n Region 185 12 NorthAmerica n Region 199 Specieswithou t an identified region 207 References 209 Indexo fbotanica l names 228 Preface The aimo f thiswor k ist ogiv e thereade r quick reference toth e regionso f diversity ofcultivate d plants.Fo r important crops,region so fdiversit y of related wild species areals opresented .Wil d species areofte nusefu l sources of genes to improve thevalu eo fcrops . -
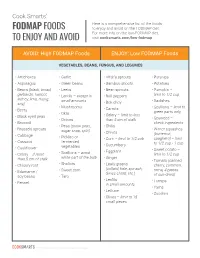
Fodmapfoods to Enjoy and Avoid
Cook Smarts’ Here is a comprehensive list of the foods FODMAP FOODS to enjoy and avoid on the FODMAP diet. For more info on the low-FODMAP diet, TO ENJOY AND AVOID visit cooksmarts.com/low-fodmap AVOID: High FODMAP Foods ENJOY: Low FODMAP Foods VEGETABLES, BEANS, FUNGUS, AND LEGUMES • Artichokes • Garlic • Alfalfa sprouts • Parsnips • Asparagus • Green beans • Bamboo shoots • Potatoes • Beans (black, broad, • Leeks • Bean sprouts • Pumpkin – garbanzo, haricot, • Lentils – except in • Bell peppers limit to 1/2 cup kidney, lima, mung, small amounts • Radishes soy) • Bok choy • Mushrooms • Scallions – limit to • Beets • Carrots • Okra green parts only • Black-eyed peas • Celery – limit to less • Onions than 5 cm of stalk • Seaweed – • Broccoli check ingredients • Peas (snow peas, • Chilis • Brussels sprouts • Winter squashes sugar snap, split) • Chives • Cabbage (butternut, • Pickled or • Corn – limit to 1/2 cob spaghetti) – limit • Cassava fermented • Cucumbers to 1/2 cup - 1 cup • Cauliflower vegetables • Eggplant • Sweet potato – • Celery – if more • Scallions – avoid limit to 1/2 cup white part of the bulb • Ginger than 5 cm of stalk • Tomato (canned, • Chicory root • Shallots • Leafy greens cherry, common, • Sweet corn (collard, kale, spinach, roma, 4 pieces • Edamame / Swiss chard, etc.) soy beans • Taro of sun-dried) • Lentils – • Turnips • Fennel in small amounts • Yams • Lettuce • Zucchini • Olives – limit to 15 small pieces © 2018 Cook Smarts | All Rights Reserved | Page 1 AVOID: High FODMAP Foods ENJOY: Low FODMAP Foods FRUITS -

ABSTRACT OH, JIYOUNG. Growth Regulator Effects on Watermelon
ABSTRACT OH, JIYOUNG. Growth regulator Effects on Watermelon Chilling Resistance, Flowering, and Fruiting. (Under the direction of Dr. Todd C. Wehner.) The watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] is classified as a tender warm season crop and is native to the Old World tropics and subtropics (Bates and Robinson, 1995). Watermelon seedlings may be exposed to temperatures between chilling and optimal for weeks before temperatures stabilize. An experiment was conducted to determine the abscisic acid effect on the watermelon chilling resistance in nine different cultivars that examined whether they were resistant or susceptible. Eighty, 160, and 320 mg/kg ABA applied 12, 24, 36 or 72 hours before chilling provided protection from chilling damage. Hybrid watermelon seeds are produced by hand pollination using alternating rows in a field for the two parental inbred lines. In some cases, a male sterile has been used to make hybrid seed production easier. However, since the male sterile is the genic type, it is necessary to check each plant to make sure it is male sterile and must be removed before pollination begins. To avoid the labor involved in removing the male flowers, we conducted a study to identify growth regulators that would convert monoecious into gynoecious. We evaluated ten different growth regulators for their effect on staminate: pistillate flower ratio in more gynoecious and androecious watermelon cultivars. AVG (aminoethylvinylglycine) with 100 mg/kg of ethrel appears to increase the percentage of pistillate nodes. However, a single treatment of AVG without ethrel had no effect on sex expression. None of the other treatments induced higher gynoecious sex expression compared with the control. -

Cultigen and Plant Spacing Effects on Plant Growth, Disease, Yield and Fruit Quality of Tomatoes
other studies (Cartia et al., 1989; Chellemi et al., 1997). Soil Heald, C. M. and A. F. Robinson. 1987. Effects of soil solarization on Rotylen- chulus reniformis in the Lower Rio Grande Valley of Texas. J. Nematol. samples measured for nematode density did not contain root- 19:93-103. knot species for any of three locations prior to planting. Thus, Jenkins, W. R. 1964. A rapid centrifugal-flotation technique for separating it is advisable to include treatments for the management of nematodes from soil. Plant Dis. Rptr. 48:692. root-knot nematodes when utilizing solarization for cucurbit McSorley, R. andj. L. Parrado. 1986. Application of soil solarization to rock- dale soils in a subtropical environment. Nematropica 16:125-140. production. Overman, A. J. 1985. Off-season land management, soil solarization and fu migation for tomato. Soil Crop Sci. Soc. Fla. Proc. 44:35-39. RistainoJ. B., K. B. Perry and R. D. Lumsden. 1991. Effect of solarization and Literature Cited Gliocladium vixens on sclerotia of Sclerotium rolfsii, soil microbiota and the incidence of southern blight of tomato. Phytopathology 81:1117-1124. Cantliffe, D. J., G. J. Hochmuth, S. J. Locascio, P. A. Stansly, C. S. Vavrina, J. Ristaino, J. B., K. B. Perry and R. D. Lumsden. 1996. Soil solarization and Glio E. Polston, D. J. Schuster, D. R. Seal, D. O. Chellemi and S. M. Olson. cladium vixens reduce the incidence of southern blight {Sclerotium rolfsii) 1995. Production of solanaceae for fresh market under field conditions: in bell pepper in the field. Biocon. Sci. and Tech. 6:583-593.